Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Irf3
Z-value: 1.96
Transcription factors associated with Irf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf3
|
ENSMUSG00000003184.16 | Irf3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf3 | mm39_v1_chr7_+_44647072_44647126 | 0.70 | 6.0e-12 | Click! |
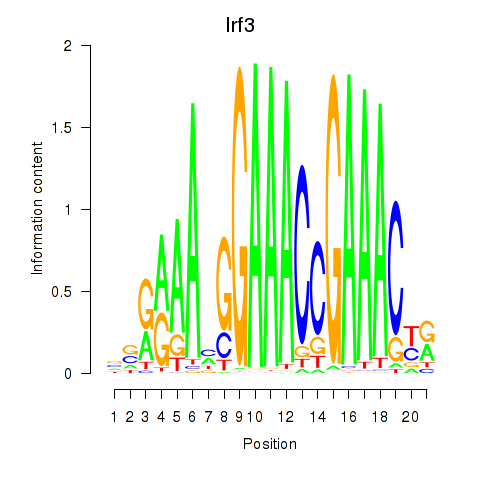
Activity profile of Irf3 motif
Sorted Z-values of Irf3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_83421333 | 26.65 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr2_-_173060647 | 19.47 |
ENSMUST00000109116.3
ENSMUST00000029018.14 |
Zbp1
|
Z-DNA binding protein 1 |
| chr15_-_77417512 | 17.50 |
ENSMUST00000062562.7
ENSMUST00000230863.2 |
Apol7c
|
apolipoprotein L 7c |
| chr11_-_48955028 | 17.33 |
ENSMUST00000046745.7
|
Tgtp2
|
T cell specific GTPase 2 |
| chr9_-_22041894 | 15.72 |
ENSMUST00000115315.3
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr7_-_103964662 | 15.37 |
ENSMUST00000106837.8
ENSMUST00000106839.9 ENSMUST00000070943.7 |
Trim12a
|
tripartite motif-containing 12A |
| chr7_-_104002534 | 15.07 |
ENSMUST00000130139.3
ENSMUST00000059037.15 |
Trim12c
|
tripartite motif-containing 12C |
| chr1_-_85247864 | 14.82 |
ENSMUST00000159582.8
ENSMUST00000161267.8 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr17_+_34406523 | 14.82 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr17_-_34406193 | 13.97 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr6_-_3399451 | 13.90 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr5_+_115034997 | 13.78 |
ENSMUST00000031542.13
ENSMUST00000146072.8 ENSMUST00000150361.2 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr7_-_103937301 | 13.65 |
ENSMUST00000098179.9
|
Trim5
|
tripartite motif-containing 5 |
| chr17_+_34406762 | 13.53 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr7_+_78563513 | 13.22 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr1_+_85528392 | 12.81 |
ENSMUST00000080204.11
|
Sp140
|
Sp140 nuclear body protein |
| chr5_-_121045568 | 12.75 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr16_-_35691914 | 12.45 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr8_-_71219299 | 12.38 |
ENSMUST00000222087.4
|
Ifi30
|
interferon gamma inducible protein 30 |
| chr7_+_78563964 | 12.37 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr11_-_48883053 | 12.25 |
ENSMUST00000059930.3
ENSMUST00000068063.4 |
Gm12185
Tgtp1
|
predicted gene 12185 T cell specific GTPase 1 |
| chr5_-_121025645 | 12.20 |
ENSMUST00000086368.12
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr18_-_32692967 | 12.00 |
ENSMUST00000174000.2
ENSMUST00000174459.2 |
Gypc
|
glycophorin C |
| chr3_+_142236086 | 11.59 |
ENSMUST00000171263.8
ENSMUST00000045097.11 |
Gbp7
|
guanylate binding protein 7 |
| chr4_-_156285247 | 11.56 |
ENSMUST00000085425.6
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr16_-_10603389 | 11.40 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chr5_+_120950690 | 10.86 |
ENSMUST00000086377.9
|
Oas1b
|
2'-5' oligoadenylate synthetase 1B |
| chr1_-_85526517 | 10.84 |
ENSMUST00000093508.7
|
Sp110
|
Sp110 nuclear body protein |
| chr7_-_140846328 | 10.55 |
ENSMUST00000106023.8
ENSMUST00000097952.9 ENSMUST00000026571.11 |
Irf7
|
interferon regulatory factor 7 |
| chr7_+_78564062 | 10.34 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chr14_-_66071412 | 9.33 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr5_-_120950570 | 9.20 |
ENSMUST00000117193.8
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr2_-_62476487 | 8.82 |
ENSMUST00000112459.4
ENSMUST00000028259.12 |
Ifih1
|
interferon induced with helicase C domain 1 |
| chr11_-_100595019 | 8.66 |
ENSMUST00000017974.13
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chrX_+_100473161 | 8.64 |
ENSMUST00000033673.7
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chr16_+_35759346 | 8.60 |
ENSMUST00000023622.13
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr17_-_36331416 | 8.37 |
ENSMUST00000174063.2
ENSMUST00000113760.10 |
H2-T24
|
histocompatibility 2, T region locus 24 |
| chr7_+_103893658 | 7.94 |
ENSMUST00000106849.9
ENSMUST00000060315.12 |
Trim34a
|
tripartite motif-containing 34A |
| chr7_+_78563184 | 7.87 |
ENSMUST00000121645.8
|
Isg20
|
interferon-stimulated protein |
| chr13_+_74787952 | 7.83 |
ENSMUST00000221822.2
ENSMUST00000221526.2 |
Erap1
|
endoplasmic reticulum aminopeptidase 1 |
| chr1_-_85087567 | 7.61 |
ENSMUST00000161675.3
ENSMUST00000160792.8 |
A530032D15Rik
|
RIKEN cDNA A530032D15Rik gene |
| chr8_-_71990085 | 7.56 |
ENSMUST00000051672.9
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chr11_+_58090382 | 7.48 |
ENSMUST00000035266.11
ENSMUST00000094169.11 ENSMUST00000168280.2 ENSMUST00000058704.9 |
Igtp
Irgm2
|
interferon gamma induced GTPase immunity-related GTPase family M member 2 |
| chr8_-_124045292 | 7.43 |
ENSMUST00000118395.2
ENSMUST00000035495.15 |
Fanca
|
Fanconi anemia, complementation group A |
| chr5_-_137145030 | 7.33 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr7_-_102214636 | 7.12 |
ENSMUST00000106913.3
ENSMUST00000033264.12 |
Trim21
|
tripartite motif-containing 21 |
| chr2_+_121978156 | 7.01 |
ENSMUST00000102476.5
|
B2m
|
beta-2 microglobulin |
| chr16_-_35759461 | 6.97 |
ENSMUST00000081933.14
ENSMUST00000114885.3 |
Dtx3l
|
deltex 3-like, E3 ubiquitin ligase |
| chr1_-_85248264 | 6.85 |
ENSMUST00000093506.12
ENSMUST00000064341.9 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr17_+_57665691 | 6.70 |
ENSMUST00000086763.13
ENSMUST00000233376.2 ENSMUST00000233840.2 ENSMUST00000232808.2 ENSMUST00000004850.8 |
Adgre1
|
adhesion G protein-coupled receptor E1 |
| chr14_+_55815999 | 6.63 |
ENSMUST00000172738.2
ENSMUST00000089619.13 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr7_+_103893672 | 6.63 |
ENSMUST00000106848.8
|
Trim34a
|
tripartite motif-containing 34A |
| chr7_-_44785480 | 6.56 |
ENSMUST00000211246.2
ENSMUST00000210197.2 |
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr7_-_100164007 | 6.49 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr14_+_55815879 | 6.02 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr14_+_55815817 | 5.86 |
ENSMUST00000174259.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr3_-_104725853 | 5.56 |
ENSMUST00000106775.8
ENSMUST00000166979.8 |
Mov10
|
Mov10 RISC complex RNA helicase |
| chr15_+_77613239 | 5.43 |
ENSMUST00000230979.2
ENSMUST00000109775.4 |
Apol9b
|
apolipoprotein L 9b |
| chr3_+_142266113 | 5.42 |
ENSMUST00000106221.8
|
Gbp3
|
guanylate binding protein 3 |
| chr6_-_125208738 | 5.37 |
ENSMUST00000043422.8
|
Tapbpl
|
TAP binding protein-like |
| chr11_+_20597149 | 5.32 |
ENSMUST00000109585.2
|
Sertad2
|
SERTA domain containing 2 |
| chr15_-_77295234 | 5.32 |
ENSMUST00000089452.6
ENSMUST00000081776.11 |
Apol9a
|
apolipoprotein L 9a |
| chr9_-_35087348 | 4.88 |
ENSMUST00000119847.2
ENSMUST00000034539.12 |
Dcps
|
decapping enzyme, scavenger |
| chr3_-_151455514 | 4.75 |
ENSMUST00000029671.9
|
Ifi44
|
interferon-induced protein 44 |
| chr5_-_120950549 | 4.71 |
ENSMUST00000125547.2
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr11_-_48762170 | 4.57 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr11_+_109253598 | 4.57 |
ENSMUST00000106702.4
ENSMUST00000020930.14 |
Gna13
|
guanine nucleotide binding protein, alpha 13 |
| chr3_-_104725581 | 4.47 |
ENSMUST00000168015.8
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr3_+_142265904 | 4.31 |
ENSMUST00000128609.8
ENSMUST00000029935.14 |
Gbp3
|
guanylate binding protein 3 |
| chr14_+_55815580 | 4.30 |
ENSMUST00000174484.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr15_-_82128538 | 4.00 |
ENSMUST00000229747.2
ENSMUST00000230408.2 |
Cenpm
|
centromere protein M |
| chr3_+_27371168 | 3.92 |
ENSMUST00000046383.12
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr2_+_93017916 | 3.88 |
ENSMUST00000090554.11
|
Trp53i11
|
transformation related protein 53 inducible protein 11 |
| chr4_-_40239778 | 3.81 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr2_-_145776934 | 3.75 |
ENSMUST00000001818.5
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr14_-_55828511 | 3.70 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr4_-_40239700 | 3.61 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr2_+_93017887 | 3.55 |
ENSMUST00000150462.8
ENSMUST00000111266.8 |
Trp53i11
|
transformation related protein 53 inducible protein 11 |
| chr7_-_44785603 | 3.44 |
ENSMUST00000209467.2
|
Gm45713
|
predicted gene 45713 |
| chr1_+_171216480 | 3.41 |
ENSMUST00000056449.9
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr3_+_142265787 | 3.35 |
ENSMUST00000199325.5
ENSMUST00000106222.9 |
Gbp3
|
guanylate binding protein 3 |
| chr7_-_44785709 | 3.02 |
ENSMUST00000211429.2
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr6_-_54949587 | 3.01 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr11_+_119283887 | 2.91 |
ENSMUST00000093902.12
ENSMUST00000131035.10 |
Rnf213
|
ring finger protein 213 |
| chr14_-_66071337 | 2.77 |
ENSMUST00000225853.2
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr16_-_19525122 | 2.75 |
ENSMUST00000081880.7
|
Lamp3
|
lysosomal-associated membrane protein 3 |
| chr6_-_38331187 | 2.52 |
ENSMUST00000114900.8
ENSMUST00000143702.5 |
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr17_+_75485791 | 2.26 |
ENSMUST00000135447.8
ENSMUST00000112516.8 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr12_+_77285770 | 2.14 |
ENSMUST00000062804.8
|
Fut8
|
fucosyltransferase 8 |
| chr10_+_128106414 | 2.11 |
ENSMUST00000085708.3
ENSMUST00000105238.10 |
Stat2
|
signal transducer and activator of transcription 2 |
| chr19_+_6096606 | 1.85 |
ENSMUST00000138532.8
ENSMUST00000129081.8 ENSMUST00000156550.8 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr6_-_38331482 | 1.78 |
ENSMUST00000031850.10
ENSMUST00000114898.3 |
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr19_-_32085532 | 1.57 |
ENSMUST00000236030.2
ENSMUST00000237104.2 ENSMUST00000236208.2 ENSMUST00000235273.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr17_+_75485906 | 1.42 |
ENSMUST00000112514.2
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr3_-_104725535 | 1.38 |
ENSMUST00000002297.12
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr10_-_87329513 | 1.34 |
ENSMUST00000020243.10
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr7_+_103064279 | 1.33 |
ENSMUST00000106892.2
|
Usp17lc
|
ubiquitin specific peptidase 17-like C |
| chr1_+_61677977 | 1.26 |
ENSMUST00000075374.10
|
Pard3b
|
par-3 family cell polarity regulator beta |
| chr17_+_21229165 | 1.23 |
ENSMUST00000232473.2
ENSMUST00000231482.2 ENSMUST00000088811.7 ENSMUST00000232354.2 ENSMUST00000232595.2 ENSMUST00000232320.2 |
Zfp160
|
zinc finger protein 160 |
| chr15_+_38976260 | 1.23 |
ENSMUST00000022909.10
|
Dcaf13
|
DDB1 and CUL4 associated factor 13 |
| chr6_-_39095144 | 1.20 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr2_-_126775136 | 1.18 |
ENSMUST00000028844.11
|
Sppl2a
|
signal peptide peptidase like 2A |
| chr13_+_42205790 | 1.15 |
ENSMUST00000220525.2
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr4_+_115595610 | 1.15 |
ENSMUST00000106524.4
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr6_+_34840057 | 1.13 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr8_-_81466126 | 1.13 |
ENSMUST00000043359.9
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr6_-_56774746 | 1.04 |
ENSMUST00000114321.2
|
Kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr3_+_27371206 | 1.03 |
ENSMUST00000174840.2
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr14_+_30438334 | 1.00 |
ENSMUST00000228006.2
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr16_-_38162174 | 0.99 |
ENSMUST00000114740.3
|
Cfap91
|
cilia and flagella associated protein 91 |
| chr11_+_72192455 | 0.97 |
ENSMUST00000151440.8
ENSMUST00000146233.8 ENSMUST00000140842.9 |
Xaf1
|
XIAP associated factor 1 |
| chr1_-_85087170 | 0.82 |
ENSMUST00000161724.8
|
A530032D15Rik
|
RIKEN cDNA A530032D15Rik gene |
| chr2_+_38888312 | 0.71 |
ENSMUST00000112872.2
|
Wdr38
|
WD repeat domain 38 |
| chr16_+_23428650 | 0.68 |
ENSMUST00000038423.6
ENSMUST00000211349.2 |
Rtp4
|
receptor transporter protein 4 |
| chr18_+_24122689 | 0.67 |
ENSMUST00000074941.8
|
Zfp35
|
zinc finger protein 35 |
| chr14_-_8457069 | 0.62 |
ENSMUST00000022257.4
|
Atxn7
|
ataxin 7 |
| chr19_+_34560922 | 0.57 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr11_+_83743746 | 0.55 |
ENSMUST00000108113.3
|
Hnf1b
|
HNF1 homeobox B |
| chr6_+_34840151 | 0.49 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr17_+_21229279 | 0.47 |
ENSMUST00000232663.2
|
Zfp160
|
zinc finger protein 160 |
| chr6_-_34294377 | 0.44 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr10_-_81332163 | 0.33 |
ENSMUST00000020463.14
|
Ncln
|
nicalin |
| chr12_-_83643964 | 0.29 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr10_-_81332147 | 0.21 |
ENSMUST00000118498.8
|
Ncln
|
nicalin |
| chr7_-_126194097 | 0.18 |
ENSMUST00000058429.6
|
Il27
|
interleukin 27 |
| chr1_-_174079627 | 0.16 |
ENSMUST00000214725.2
|
Olfr419
|
olfactory receptor 419 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 28.4 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 7.3 | 43.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 6.7 | 26.6 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 5.4 | 5.4 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 3.5 | 7.0 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 3.1 | 12.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 2.9 | 11.4 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 2.7 | 16.2 | GO:0035547 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 2.5 | 7.5 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 2.4 | 12.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 2.0 | 15.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 1.9 | 9.6 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 1.8 | 10.6 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 1.6 | 11.4 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 1.6 | 4.9 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 1.6 | 6.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.4 | 11.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.4 | 8.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 1.4 | 13.9 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 1.2 | 19.5 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 1.1 | 12.4 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 1.0 | 26.5 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 1.0 | 47.8 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 1.0 | 8.6 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.8 | 7.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.7 | 7.4 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.7 | 8.5 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.7 | 8.6 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.6 | 9.4 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.6 | 11.6 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.5 | 2.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.5 | 4.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.4 | 1.3 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) olfactory pit development(GO:0060166) |
| 0.4 | 0.4 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 0.4 | 3.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.3 | 22.3 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.3 | 4.3 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.3 | 2.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.3 | 1.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 2.7 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.3 | 2.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 57.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 0.6 | GO:0061296 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 0.7 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 9.4 | GO:0048525 | negative regulation of viral process(GO:0048525) |
| 0.1 | 12.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 4.8 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 3.7 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 1.8 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 3.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.2 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 6.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 5.3 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 1.0 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 8.4 | GO:0045087 | innate immune response(GO:0045087) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 22.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 3.5 | 28.4 | GO:0042825 | TAP complex(GO:0042825) |
| 2.9 | 29.0 | GO:1990462 | omegasome(GO:1990462) |
| 2.3 | 13.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 2.0 | 32.2 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 1.2 | 3.7 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.9 | 2.7 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.9 | 8.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.6 | 12.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.5 | 30.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.5 | 7.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 7.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 1.8 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 3.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.3 | 9.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.3 | 4.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 25.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 1.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 2.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 7.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 7.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 4.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 12.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 9.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 6.5 | GO:0005930 | axoneme(GO:0005930) |
| 0.1 | 30.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 7.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 28.7 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 7.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 4.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 5.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 6.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 28.4 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 8.9 | 26.6 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 8.8 | 43.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 6.5 | 19.5 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 4.4 | 52.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 2.4 | 26.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.2 | 28.7 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 1.1 | 4.6 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 1.1 | 12.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.9 | 3.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.9 | 26.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.9 | 10.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.9 | 15.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.8 | 7.8 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.7 | 11.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.6 | 14.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 7.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 12.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.3 | 4.9 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.3 | 11.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 8.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 1.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 35.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 2.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 47.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.8 | GO:1990381 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 4.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 9.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 9.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 9.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 6.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 26.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 5.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 12.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 26.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 8.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 11.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 4.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 2.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 7.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 12.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 13.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 28.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 7.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 10.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 3.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.9 | 36.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 1.2 | 26.8 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 1.0 | 70.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.7 | 19.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.7 | 37.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.5 | 26.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 4.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 11.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.3 | 4.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 11.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.3 | 7.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 3.0 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.2 | 8.7 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.2 | 14.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 4.6 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 2.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 1.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 4.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 7.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |