Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
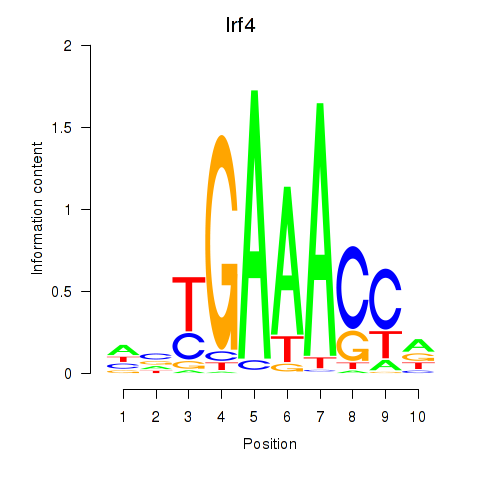
Results for Irf4
Z-value: 1.21
Transcription factors associated with Irf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf4
|
ENSMUSG00000021356.11 | Irf4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf4 | mm39_v1_chr13_+_30933209_30933290 | 0.34 | 3.1e-03 | Click! |
Activity profile of Irf4 motif
Sorted Z-values of Irf4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_36293588 | 11.85 |
ENSMUST00000060128.7
|
Cldn23
|
claudin 23 |
| chr4_-_89229256 | 10.98 |
ENSMUST00000097981.6
|
Cdkn2b
|
cyclin dependent kinase inhibitor 2B |
| chr11_+_69857722 | 10.36 |
ENSMUST00000151515.2
|
Cldn7
|
claudin 7 |
| chr9_+_44953723 | 8.35 |
ENSMUST00000034600.5
|
Mpzl2
|
myelin protein zero-like 2 |
| chr3_+_3699205 | 8.04 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr19_+_56385531 | 7.55 |
ENSMUST00000026062.10
|
Casp7
|
caspase 7 |
| chr13_-_95661726 | 7.37 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr13_-_100689212 | 7.28 |
ENSMUST00000022140.12
|
Ocln
|
occludin |
| chr13_-_100753419 | 7.28 |
ENSMUST00000168772.2
ENSMUST00000163163.9 ENSMUST00000022137.14 |
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr11_+_115353290 | 7.27 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr13_-_100689105 | 7.21 |
ENSMUST00000159459.8
|
Ocln
|
occludin |
| chr3_+_3573084 | 7.10 |
ENSMUST00000108393.8
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr17_-_7050145 | 7.05 |
ENSMUST00000064234.7
|
Ezr
|
ezrin |
| chr9_-_65330231 | 6.57 |
ENSMUST00000065894.7
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr6_-_129077867 | 6.53 |
ENSMUST00000032258.8
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr8_+_46338498 | 6.51 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr7_+_79836581 | 6.47 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr8_+_46338557 | 6.16 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr3_-_30753979 | 6.07 |
ENSMUST00000126658.8
ENSMUST00000108263.2 |
Lrrc31
|
leucine rich repeat containing 31 |
| chr17_+_35481702 | 6.06 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr4_-_132990362 | 5.85 |
ENSMUST00000105908.10
ENSMUST00000030674.8 |
Sytl1
|
synaptotagmin-like 1 |
| chr4_-_46536088 | 5.74 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr2_-_173060647 | 5.61 |
ENSMUST00000109116.3
ENSMUST00000029018.14 |
Zbp1
|
Z-DNA binding protein 1 |
| chr13_-_100753181 | 5.56 |
ENSMUST00000225754.2
|
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chrX_+_138464065 | 5.56 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr11_+_62711057 | 5.52 |
ENSMUST00000055006.12
ENSMUST00000072639.10 |
Trim16
|
tripartite motif-containing 16 |
| chr16_-_55659194 | 5.51 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr7_-_18757461 | 5.42 |
ENSMUST00000036018.6
|
Foxa3
|
forkhead box A3 |
| chr7_-_4606104 | 5.29 |
ENSMUST00000049113.14
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr4_-_129132963 | 4.93 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080 |
| chr17_-_29134288 | 4.88 |
ENSMUST00000062357.6
|
Bnip5
|
BCL2 interacting protein 5 |
| chr14_-_54646917 | 4.73 |
ENSMUST00000000984.9
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr3_+_27371168 | 4.57 |
ENSMUST00000046383.12
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr11_+_82006001 | 4.57 |
ENSMUST00000009329.3
|
Ccl8
|
chemokine (C-C motif) ligand 8 |
| chr19_-_57185928 | 4.49 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_-_140846328 | 4.33 |
ENSMUST00000106023.8
ENSMUST00000097952.9 ENSMUST00000026571.11 |
Irf7
|
interferon regulatory factor 7 |
| chr11_+_62711295 | 4.28 |
ENSMUST00000108703.2
|
Trim16
|
tripartite motif-containing 16 |
| chr12_+_119354110 | 4.24 |
ENSMUST00000222058.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr10_-_111833138 | 4.21 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr19_-_57185808 | 4.09 |
ENSMUST00000111546.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr5_-_104169696 | 4.06 |
ENSMUST00000119025.2
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr11_-_3489228 | 4.06 |
ENSMUST00000075118.10
ENSMUST00000136243.2 ENSMUST00000020721.15 ENSMUST00000170588.8 |
Smtn
|
smoothelin |
| chr19_-_57185988 | 3.99 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr11_-_101062111 | 3.98 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr19_-_24202344 | 3.87 |
ENSMUST00000099558.5
ENSMUST00000232956.2 |
Tjp2
|
tight junction protein 2 |
| chr6_-_70313491 | 3.84 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr12_+_111383864 | 3.67 |
ENSMUST00000220537.2
ENSMUST00000223050.2 ENSMUST00000072646.8 ENSMUST00000223431.2 ENSMUST00000221144.2 ENSMUST00000222437.2 |
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr3_+_95434093 | 3.64 |
ENSMUST00000015667.9
ENSMUST00000116304.3 |
Ctss
|
cathepsin S |
| chr16_-_43484494 | 3.62 |
ENSMUST00000096065.6
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr3_+_27371206 | 3.59 |
ENSMUST00000174840.2
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr11_+_48977852 | 3.59 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr19_-_57185861 | 3.57 |
ENSMUST00000111550.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_-_89176294 | 3.56 |
ENSMUST00000207932.2
|
Prss23
|
protease, serine 23 |
| chr5_-_104169785 | 3.51 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr6_-_87312743 | 3.43 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr11_+_48977888 | 3.35 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr10_+_82696135 | 3.34 |
ENSMUST00000219442.3
|
Txnrd1
|
thioredoxin reductase 1 |
| chr16_+_35759346 | 3.25 |
ENSMUST00000023622.13
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr2_+_38401826 | 3.10 |
ENSMUST00000112895.8
|
Nek6
|
NIMA (never in mitosis gene a)-related expressed kinase 6 |
| chr9_-_116004386 | 2.96 |
ENSMUST00000035014.8
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr19_-_34618135 | 2.95 |
ENSMUST00000087357.5
|
Ifit1bl2
|
interferon induced protein with tetratricopeptide repeats 1B like 2 |
| chr6_-_87312681 | 2.94 |
ENSMUST00000204805.3
|
Antxr1
|
anthrax toxin receptor 1 |
| chr10_-_53952686 | 2.88 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr10_+_69048464 | 2.77 |
ENSMUST00000020101.12
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr15_-_58187556 | 2.73 |
ENSMUST00000022985.2
|
Klhl38
|
kelch-like 38 |
| chr10_+_94411119 | 2.73 |
ENSMUST00000121471.8
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr2_+_127967951 | 2.72 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr1_+_182236728 | 2.66 |
ENSMUST00000117245.2
|
Trp53bp2
|
transformation related protein 53 binding protein 2 |
| chrX_-_9335525 | 2.65 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr1_-_163141278 | 2.65 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr1_-_174749379 | 2.63 |
ENSMUST00000055294.4
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chrX_+_158086253 | 2.63 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr19_+_34618271 | 2.60 |
ENSMUST00000102824.4
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr16_-_35759461 | 2.58 |
ENSMUST00000081933.14
ENSMUST00000114885.3 |
Dtx3l
|
deltex 3-like, E3 ubiquitin ligase |
| chr1_+_57884693 | 2.57 |
ENSMUST00000169772.3
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr11_+_43365103 | 2.56 |
ENSMUST00000173002.8
ENSMUST00000057679.10 |
C1qtnf2
|
C1q and tumor necrosis factor related protein 2 |
| chr8_-_25086976 | 2.49 |
ENSMUST00000033956.7
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chrX_-_107877909 | 2.47 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr11_-_101442663 | 2.42 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr6_+_53264255 | 2.41 |
ENSMUST00000203528.3
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr1_+_16758629 | 2.39 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr3_+_89328926 | 2.36 |
ENSMUST00000094378.10
ENSMUST00000137793.2 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr1_-_156766381 | 2.32 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr3_+_90444613 | 2.31 |
ENSMUST00000107335.2
|
S100a16
|
S100 calcium binding protein A16 |
| chr7_-_4448631 | 2.29 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr2_+_38401655 | 2.26 |
ENSMUST00000054234.10
ENSMUST00000112902.8 |
Nek6
|
NIMA (never in mitosis gene a)-related expressed kinase 6 |
| chr1_-_164763091 | 2.26 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr9_+_5345405 | 2.26 |
ENSMUST00000027009.11
|
Casp12
|
caspase 12 |
| chr3_+_90444537 | 2.25 |
ENSMUST00000098911.10
|
S100a16
|
S100 calcium binding protein A16 |
| chr9_+_5345450 | 2.25 |
ENSMUST00000151332.2
|
Casp12
|
caspase 12 |
| chr1_-_156766351 | 2.24 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr12_-_114073050 | 2.21 |
ENSMUST00000103472.4
|
Ighv9-2
|
immunoglobulin heavy variable V9-2 |
| chrX_-_105884178 | 2.20 |
ENSMUST00000062010.10
|
Rtl3
|
retrotransposon Gag like 3 |
| chr4_-_156285247 | 2.12 |
ENSMUST00000085425.6
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr7_-_65020655 | 2.05 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr10_-_12424623 | 2.01 |
ENSMUST00000219003.2
|
Utrn
|
utrophin |
| chr14_-_30348153 | 2.01 |
ENSMUST00000112211.9
ENSMUST00000112210.11 |
Prkcd
|
protein kinase C, delta |
| chr14_+_14475188 | 2.00 |
ENSMUST00000026315.8
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr3_-_142101418 | 2.00 |
ENSMUST00000029941.16
ENSMUST00000058626.9 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr10_+_21758083 | 1.96 |
ENSMUST00000120509.8
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr1_-_192883642 | 1.91 |
ENSMUST00000192020.6
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr8_+_79236051 | 1.83 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr15_-_76127600 | 1.83 |
ENSMUST00000165738.8
ENSMUST00000075689.7 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr17_-_35081129 | 1.82 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr19_-_4976844 | 1.81 |
ENSMUST00000236496.2
|
Dpp3
|
dipeptidylpeptidase 3 |
| chr7_-_80037622 | 1.81 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chr1_+_61677977 | 1.77 |
ENSMUST00000075374.10
|
Pard3b
|
par-3 family cell polarity regulator beta |
| chr17_-_35081456 | 1.71 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr7_+_126895531 | 1.70 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr13_-_64645606 | 1.68 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr1_+_160898283 | 1.67 |
ENSMUST00000028035.14
ENSMUST00000111620.10 ENSMUST00000111618.8 |
Cenpl
|
centromere protein L |
| chr14_+_120513076 | 1.62 |
ENSMUST00000088419.13
|
Mbnl2
|
muscleblind like splicing factor 2 |
| chr5_-_99391073 | 1.55 |
ENSMUST00000166484.2
|
Rasgef1b
|
RasGEF domain family, member 1B |
| chr10_+_18283405 | 1.53 |
ENSMUST00000037341.14
|
Nhsl1
|
NHS-like 1 |
| chr12_-_118930130 | 1.50 |
ENSMUST00000035515.5
|
Abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr10_+_69048506 | 1.48 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr10_+_28544356 | 1.47 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr7_+_126895463 | 1.41 |
ENSMUST00000106306.9
ENSMUST00000120857.8 |
Itgal
|
integrin alpha L |
| chr14_+_54436247 | 1.37 |
ENSMUST00000103720.2
|
Traj21
|
T cell receptor alpha joining 21 |
| chr15_-_101651534 | 1.36 |
ENSMUST00000023710.6
|
Krt71
|
keratin 71 |
| chr11_+_78136569 | 1.35 |
ENSMUST00000002133.9
|
Sdf2
|
stromal cell derived factor 2 |
| chr6_-_39095144 | 1.32 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr8_+_105897300 | 1.32 |
ENSMUST00000052209.9
ENSMUST00000109392.9 ENSMUST00000109395.8 |
Cbfb
|
core binding factor beta |
| chr14_+_55815879 | 1.31 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr2_-_131016573 | 1.28 |
ENSMUST00000127987.2
|
Spef1
|
sperm flagellar 1 |
| chr19_-_3955230 | 1.27 |
ENSMUST00000145791.8
|
Tcirg1
|
T cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3 |
| chr3_-_151953894 | 1.27 |
ENSMUST00000196529.5
|
Nexn
|
nexilin |
| chr4_+_114857348 | 1.25 |
ENSMUST00000030490.13
|
Stil
|
Scl/Tal1 interrupting locus |
| chr7_+_35285657 | 1.22 |
ENSMUST00000040844.16
ENSMUST00000188906.7 ENSMUST00000186245.7 ENSMUST00000190503.7 |
Ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr1_+_52158693 | 1.20 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr14_+_55815580 | 1.19 |
ENSMUST00000174484.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr7_+_126895423 | 1.18 |
ENSMUST00000117762.8
|
Itgal
|
integrin alpha L |
| chr14_+_55815817 | 1.15 |
ENSMUST00000174259.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr3_+_60436570 | 1.14 |
ENSMUST00000192607.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr3_-_57202301 | 1.14 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr1_-_163141230 | 1.11 |
ENSMUST00000174397.3
ENSMUST00000075805.13 |
Prrx1
|
paired related homeobox 1 |
| chr4_-_123033721 | 1.09 |
ENSMUST00000030404.5
|
Ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr1_-_156766957 | 1.05 |
ENSMUST00000171292.8
ENSMUST00000063199.13 ENSMUST00000027886.14 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr1_+_16758731 | 1.02 |
ENSMUST00000190366.2
|
Ly96
|
lymphocyte antigen 96 |
| chr14_+_55815999 | 1.02 |
ENSMUST00000172738.2
ENSMUST00000089619.13 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr9_-_35179042 | 0.99 |
ENSMUST00000217306.2
ENSMUST00000125087.2 ENSMUST00000121564.8 ENSMUST00000063782.12 ENSMUST00000059057.14 |
Fam118b
|
family with sequence similarity 118, member B |
| chr13_-_113182891 | 0.97 |
ENSMUST00000231962.2
ENSMUST00000022282.6 |
Gpx8
|
glutathione peroxidase 8 (putative) |
| chrM_+_8603 | 0.97 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr13_+_38009981 | 0.97 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr1_+_52158599 | 0.97 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr14_+_34097474 | 0.96 |
ENSMUST00000227130.2
|
Mmrn2
|
multimerin 2 |
| chr15_-_66432938 | 0.95 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr4_-_19922599 | 0.93 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr6_-_48422447 | 0.91 |
ENSMUST00000114564.8
|
Zfp467
|
zinc finger protein 467 |
| chr4_-_107035479 | 0.91 |
ENSMUST00000058585.14
|
Tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr3_+_92325386 | 0.90 |
ENSMUST00000029533.3
|
Sprr2j-ps
|
small proline-rich protein 2J, pseudogene |
| chr5_-_92653377 | 0.89 |
ENSMUST00000031377.9
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr14_+_54436898 | 0.86 |
ENSMUST00000103721.3
|
Traj20
|
T cell receptor alpha joining 20 |
| chr4_+_114857370 | 0.84 |
ENSMUST00000129957.8
|
Stil
|
Scl/Tal1 interrupting locus |
| chr19_+_5618096 | 0.84 |
ENSMUST00000096318.4
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chrX_-_100266032 | 0.84 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr6_+_67243967 | 0.84 |
ENSMUST00000203436.3
ENSMUST00000205106.3 ENSMUST00000204293.3 ENSMUST00000203077.3 ENSMUST00000204294.3 |
Serbp1
|
serpine1 mRNA binding protein 1 |
| chr11_+_119283887 | 0.82 |
ENSMUST00000093902.12
ENSMUST00000131035.10 |
Rnf213
|
ring finger protein 213 |
| chr13_-_19579961 | 0.81 |
ENSMUST00000039694.13
|
Stard3nl
|
STARD3 N-terminal like |
| chr9_-_117080869 | 0.78 |
ENSMUST00000172564.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr14_+_120513106 | 0.78 |
ENSMUST00000227012.2
ENSMUST00000167459.3 |
Mbnl2
|
muscleblind like splicing factor 2 |
| chr1_-_75294234 | 0.76 |
ENSMUST00000066668.14
ENSMUST00000185797.7 |
Dnpep
|
aspartyl aminopeptidase |
| chr7_-_80037688 | 0.76 |
ENSMUST00000080932.8
|
Fes
|
feline sarcoma oncogene |
| chr2_-_152673032 | 0.75 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr10_-_52071340 | 0.74 |
ENSMUST00000020045.10
|
Ros1
|
Ros1 proto-oncogene |
| chr8_-_55340024 | 0.72 |
ENSMUST00000176866.8
|
Wdr17
|
WD repeat domain 17 |
| chr1_-_184464810 | 0.71 |
ENSMUST00000048572.7
|
Hlx
|
H2.0-like homeobox |
| chr17_+_35780977 | 0.70 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr13_-_19579898 | 0.68 |
ENSMUST00000197565.3
ENSMUST00000221380.2 ENSMUST00000200323.3 ENSMUST00000199924.2 ENSMUST00000222869.2 |
Stard3nl
|
STARD3 N-terminal like |
| chr11_+_87938626 | 0.68 |
ENSMUST00000107920.10
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr19_+_46611826 | 0.67 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr1_+_44158111 | 0.60 |
ENSMUST00000155917.8
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr11_+_87938519 | 0.59 |
ENSMUST00000079866.11
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr17_-_30831576 | 0.59 |
ENSMUST00000235171.2
ENSMUST00000236335.2 ENSMUST00000167624.2 |
Glo1
|
glyoxalase 1 |
| chr8_-_106665060 | 0.58 |
ENSMUST00000034369.10
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10 |
| chr5_+_147206769 | 0.53 |
ENSMUST00000085591.7
|
Pdx1
|
pancreatic and duodenal homeobox 1 |
| chr1_-_63726062 | 0.52 |
ENSMUST00000090313.5
|
Dytn
|
dystrotelin |
| chr8_-_46577183 | 0.45 |
ENSMUST00000170416.8
|
Snx25
|
sorting nexin 25 |
| chr11_+_46701619 | 0.45 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr8_+_62381115 | 0.43 |
ENSMUST00000154398.8
ENSMUST00000156980.8 ENSMUST00000093485.3 ENSMUST00000070631.15 |
Ddx60
|
DExD/H box helicase 60 |
| chr8_-_85500998 | 0.43 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr1_+_52158721 | 0.41 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr15_+_102391614 | 0.38 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr1_-_75293942 | 0.38 |
ENSMUST00000187075.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr5_-_92496730 | 0.36 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr12_-_113542610 | 0.33 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr2_-_9883391 | 0.32 |
ENSMUST00000102976.4
|
Gata3
|
GATA binding protein 3 |
| chr11_+_72580823 | 0.31 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr13_-_81718759 | 0.31 |
ENSMUST00000109565.9
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr2_+_90770742 | 0.28 |
ENSMUST00000005643.14
ENSMUST00000111451.10 ENSMUST00000177642.8 ENSMUST00000068726.13 ENSMUST00000068747.14 |
Celf1
|
CUGBP, Elav-like family member 1 |
| chr19_+_41017714 | 0.26 |
ENSMUST00000051806.12
ENSMUST00000112200.3 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr16_+_43056218 | 0.22 |
ENSMUST00000146708.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr14_-_20784625 | 0.21 |
ENSMUST00000223679.2
|
Ndst2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr9_-_110886306 | 0.21 |
ENSMUST00000195968.2
ENSMUST00000111888.3 |
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr16_+_65317389 | 0.20 |
ENSMUST00000176330.8
ENSMUST00000004964.15 ENSMUST00000176038.8 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr10_+_128542120 | 0.20 |
ENSMUST00000054125.9
|
Pmel
|
premelanosome protein |
| chr2_+_91480513 | 0.19 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chrX_-_162332673 | 0.16 |
ENSMUST00000033730.3
|
Grpr
|
gastrin releasing peptide receptor |
| chr10_-_107555840 | 0.14 |
ENSMUST00000050702.9
|
Ptprq
|
protein tyrosine phosphatase, receptor type, Q |
| chr19_+_5618029 | 0.13 |
ENSMUST00000235575.2
ENSMUST00000235542.2 |
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 1.9 | 7.6 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 1.8 | 7.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.5 | 7.5 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 1.4 | 5.4 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 1.3 | 5.2 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 1.2 | 3.6 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 1.2 | 6.1 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 1.1 | 6.4 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.9 | 2.6 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.9 | 2.6 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.8 | 2.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.8 | 14.5 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.8 | 9.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.7 | 1.5 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.7 | 4.3 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.7 | 2.7 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.6 | 4.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.6 | 5.6 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.6 | 3.9 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.5 | 11.0 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.5 | 6.6 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.5 | 2.0 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.5 | 3.4 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.5 | 2.4 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.5 | 3.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.5 | 4.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.5 | 10.4 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.5 | 1.8 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein poly-ADP-ribosylation(GO:0070212) |
| 0.4 | 1.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.4 | 2.4 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.3 | 2.7 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.3 | 5.6 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.3 | 2.0 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.3 | 2.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.3 | 0.8 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 2.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 3.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.3 | 4.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 14.9 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.2 | 3.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 3.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 3.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.2 | 8.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 1.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.2 | 8.1 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.2 | 0.7 | GO:0060154 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) response to cycloheximide(GO:0046898) cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.2 | 5.7 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 2.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.2 | 2.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 1.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.7 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 1.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 4.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 4.7 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.1 | 1.3 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 2.0 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.7 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 2.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 2.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 3.6 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 | 4.6 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 5.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 14.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 2.6 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.1 | 0.3 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.5 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 6.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 2.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 5.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 1.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.4 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.1 | 0.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.8 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 8.4 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 1.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 2.7 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.1 | 1.0 | GO:0090051 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 | 0.6 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 3.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 11.5 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 1.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.9 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 1.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 8.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 2.6 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 3.7 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 2.0 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.0 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.1 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 2.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 1.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.2 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 2.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 1.4 | 12.8 | GO:0033010 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.9 | 4.3 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.8 | 2.4 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.8 | 26.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.8 | 4.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 6.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 3.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 6.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 7.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 2.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 3.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 2.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 0.9 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 7.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 2.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 3.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 15.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 2.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 2.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 7.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 12.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 9.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 5.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 2.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 15.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 6.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 45.5 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 1.8 | 7.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.0 | 3.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.9 | 4.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.8 | 3.3 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.8 | 2.5 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.8 | 9.8 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.6 | 12.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.6 | 11.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.6 | 3.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.5 | 2.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.4 | 7.6 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.4 | 4.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 5.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 2.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.4 | 6.8 | GO:0046977 | TAP binding(GO:0046977) |
| 0.3 | 7.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 5.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 2.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 2.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 9.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 4.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 2.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 5.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 6.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 13.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 2.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 8.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 2.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 15.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 1.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.8 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 7.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 3.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 2.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 2.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 2.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 6.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.2 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 3.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 2.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 5.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 5.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 2.0 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 7.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 4.9 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 18.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 3.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 11.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 7.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 8.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 4.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 3.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 6.8 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 10.1 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 6.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 2.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 9.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 7.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 3.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 8.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 19.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 17.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 5.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 6.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 9.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 2.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 5.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.6 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.9 | 20.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.6 | 21.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.5 | 16.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.5 | 8.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 7.8 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.3 | 11.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 5.6 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.3 | 3.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 7.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 10.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 9.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 3.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 2.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 2.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 4.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 4.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 5.3 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 2.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 4.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 2.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 4.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 2.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |