Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Irf5_Irf6
Z-value: 1.49
Transcription factors associated with Irf5_Irf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
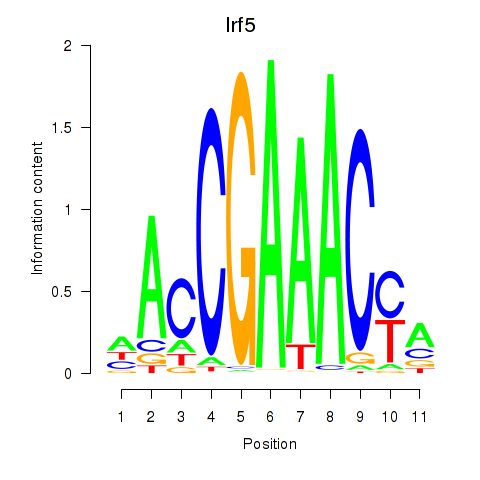
Irf5
|
ENSMUSG00000029771.13 | Irf5 |
|
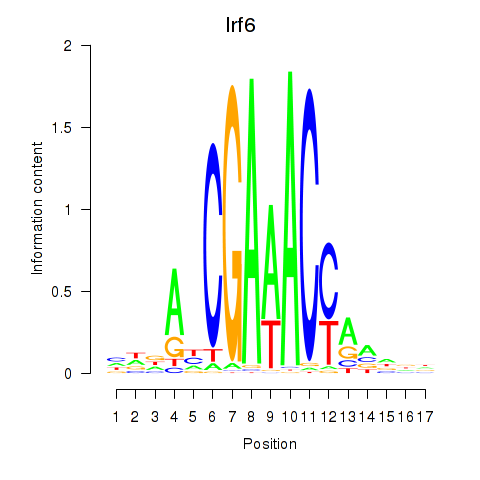
Irf6
|
ENSMUSG00000026638.16 | Irf6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf6 | mm39_v1_chr1_+_192835414_192835516 | 0.82 | 2.8e-18 | Click! |
| Irf5 | mm39_v1_chr6_+_29526679_29526723 | -0.10 | 4.2e-01 | Click! |
Activity profile of Irf5_Irf6 motif
Sorted Z-values of Irf5_Irf6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf5_Irf6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_36293588 | 36.31 |
ENSMUST00000060128.7
|
Cldn23
|
claudin 23 |
| chr9_+_44953723 | 20.95 |
ENSMUST00000034600.5
|
Mpzl2
|
myelin protein zero-like 2 |
| chr11_+_69857722 | 15.45 |
ENSMUST00000151515.2
|
Cldn7
|
claudin 7 |
| chr17_-_35081129 | 14.41 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr13_-_100689105 | 13.79 |
ENSMUST00000159459.8
|
Ocln
|
occludin |
| chr13_-_100689212 | 13.30 |
ENSMUST00000022140.12
|
Ocln
|
occludin |
| chr11_+_88890202 | 12.65 |
ENSMUST00000100627.9
ENSMUST00000107896.10 ENSMUST00000000284.7 |
Trim25
|
tripartite motif-containing 25 |
| chr17_-_35081456 | 12.35 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr1_+_130754413 | 10.48 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr19_+_5618096 | 10.17 |
ENSMUST00000096318.4
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr13_-_100688949 | 9.78 |
ENSMUST00000159515.2
ENSMUST00000160859.8 ENSMUST00000069756.11 |
Ocln
|
occludin |
| chr3_-_142101418 | 9.76 |
ENSMUST00000029941.16
ENSMUST00000058626.9 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr17_-_7050145 | 9.14 |
ENSMUST00000064234.7
|
Ezr
|
ezrin |
| chr6_+_113448388 | 8.42 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr4_-_46536088 | 6.94 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr7_-_140846328 | 6.81 |
ENSMUST00000106023.8
ENSMUST00000097952.9 ENSMUST00000026571.11 |
Irf7
|
interferon regulatory factor 7 |
| chr19_+_5618029 | 6.72 |
ENSMUST00000235575.2
ENSMUST00000235542.2 |
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr6_-_39095144 | 6.66 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr17_-_34506744 | 6.29 |
ENSMUST00000174751.2
ENSMUST00000040655.14 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr11_-_78875657 | 5.90 |
ENSMUST00000073001.5
|
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr11_-_78875689 | 5.82 |
ENSMUST00000108269.10
ENSMUST00000108268.10 |
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr17_+_31652029 | 5.76 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr16_-_35691914 | 5.59 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr19_-_5610628 | 5.29 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr17_+_31652073 | 5.25 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr17_-_79190002 | 5.18 |
ENSMUST00000024884.5
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr11_+_101473062 | 5.05 |
ENSMUST00000039581.14
ENSMUST00000100403.9 ENSMUST00000107194.8 ENSMUST00000128614.2 |
Tmem106a
|
transmembrane protein 106A |
| chr2_-_69416365 | 4.83 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr1_-_75294234 | 4.65 |
ENSMUST00000066668.14
ENSMUST00000185797.7 |
Dnpep
|
aspartyl aminopeptidase |
| chr6_+_4505493 | 4.29 |
ENSMUST00000031668.10
|
Col1a2
|
collagen, type I, alpha 2 |
| chr16_+_35758836 | 4.22 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr6_-_38331187 | 4.18 |
ENSMUST00000114900.8
ENSMUST00000143702.5 |
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr12_+_119354110 | 3.88 |
ENSMUST00000222058.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr7_-_65020655 | 3.82 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr5_-_91550853 | 3.80 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr16_-_55659194 | 3.70 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr14_+_78141679 | 3.68 |
ENSMUST00000022591.16
ENSMUST00000169978.2 ENSMUST00000227903.2 |
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr3_-_142101339 | 3.66 |
ENSMUST00000198381.5
ENSMUST00000090134.12 ENSMUST00000196908.5 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr5_+_92957231 | 3.57 |
ENSMUST00000113054.9
|
Shroom3
|
shroom family member 3 |
| chr7_+_128125339 | 3.26 |
ENSMUST00000033136.9
|
Bag3
|
BCL2-associated athanogene 3 |
| chr11_-_86091970 | 3.22 |
ENSMUST00000044423.4
|
Brip1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chr12_-_32000169 | 3.20 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr1_+_182236728 | 3.17 |
ENSMUST00000117245.2
|
Trp53bp2
|
transformation related protein 53 binding protein 2 |
| chr1_-_75293942 | 3.05 |
ENSMUST00000187075.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr15_-_76127600 | 3.04 |
ENSMUST00000165738.8
ENSMUST00000075689.7 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr7_-_26928029 | 2.94 |
ENSMUST00000003850.8
|
Itpkc
|
inositol 1,4,5-trisphosphate 3-kinase C |
| chr17_+_35780977 | 2.90 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr6_-_70313491 | 2.87 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr1_-_75293997 | 2.78 |
ENSMUST00000189282.3
ENSMUST00000191254.7 |
Dnpep
|
aspartyl aminopeptidase |
| chr8_+_62381115 | 2.56 |
ENSMUST00000154398.8
ENSMUST00000156980.8 ENSMUST00000093485.3 ENSMUST00000070631.15 |
Ddx60
|
DExD/H box helicase 60 |
| chr19_+_44744484 | 2.51 |
ENSMUST00000174490.9
|
Pax2
|
paired box 2 |
| chr1_-_75293447 | 2.47 |
ENSMUST00000189551.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr16_-_35759461 | 2.43 |
ENSMUST00000081933.14
ENSMUST00000114885.3 |
Dtx3l
|
deltex 3-like, E3 ubiquitin ligase |
| chr12_-_32000209 | 2.35 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr1_-_75293637 | 2.32 |
ENSMUST00000185419.7
ENSMUST00000187000.7 |
Dnpep
|
aspartyl aminopeptidase |
| chr17_+_23898633 | 2.24 |
ENSMUST00000233364.2
|
Cldn6
|
claudin 6 |
| chr17_-_32408431 | 2.24 |
ENSMUST00000087721.10
ENSMUST00000162117.3 |
Ephx3
|
epoxide hydrolase 3 |
| chr1_-_75293793 | 2.15 |
ENSMUST00000187836.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr4_-_40239700 | 2.13 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr15_+_96185399 | 2.13 |
ENSMUST00000134985.9
ENSMUST00000096250.5 |
Arid2
|
AT rich interactive domain 2 (ARID, RFX-like) |
| chr1_-_105284383 | 2.11 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr14_-_20784625 | 1.85 |
ENSMUST00000223679.2
|
Ndst2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr16_+_11131676 | 1.83 |
ENSMUST00000023140.6
|
Tnfrsf17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr17_+_23898223 | 1.78 |
ENSMUST00000024699.4
ENSMUST00000232719.2 |
Cldn6
|
claudin 6 |
| chr4_-_40239778 | 1.72 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr7_-_80037622 | 1.64 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chr13_-_59970383 | 1.63 |
ENSMUST00000225987.2
|
Tut7
|
terminal uridylyl transferase 7 |
| chr1_-_163141278 | 1.62 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr9_+_56979307 | 1.59 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr6_-_38331482 | 1.59 |
ENSMUST00000031850.10
ENSMUST00000114898.3 |
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr1_+_52158693 | 1.58 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr1_-_105284407 | 1.52 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr6_-_13607963 | 1.52 |
ENSMUST00000031554.9
ENSMUST00000149123.3 |
Tmem168
|
transmembrane protein 168 |
| chr4_-_116508842 | 1.52 |
ENSMUST00000030455.15
|
Akr1a1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr1_-_75293589 | 1.51 |
ENSMUST00000113605.10
|
Dnpep
|
aspartyl aminopeptidase |
| chr8_+_72943455 | 1.36 |
ENSMUST00000072097.14
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr5_-_99391073 | 1.30 |
ENSMUST00000166484.2
|
Rasgef1b
|
RasGEF domain family, member 1B |
| chr3_+_135144859 | 1.26 |
ENSMUST00000197539.5
ENSMUST00000197134.3 |
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_146855880 | 1.23 |
ENSMUST00000111622.2
ENSMUST00000036592.15 |
1700034J05Rik
|
RIKEN cDNA 1700034J05 gene |
| chr11_+_72580823 | 1.13 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr12_-_73160181 | 1.12 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chr1_+_175459559 | 1.10 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr3_+_135144481 | 1.09 |
ENSMUST00000199582.5
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_84564623 | 1.06 |
ENSMUST00000205228.3
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr5_-_100126707 | 1.06 |
ENSMUST00000170912.2
|
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr5_-_100126773 | 1.01 |
ENSMUST00000112939.10
ENSMUST00000171786.8 ENSMUST00000072750.13 ENSMUST00000019128.15 ENSMUST00000172361.8 |
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr1_+_52158599 | 0.93 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr12_-_26506422 | 0.91 |
ENSMUST00000020970.10
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr1_-_184464810 | 0.87 |
ENSMUST00000048572.7
|
Hlx
|
H2.0-like homeobox |
| chr6_-_28449250 | 0.84 |
ENSMUST00000164519.9
ENSMUST00000171089.9 ENSMUST00000031718.14 |
Pax4
|
paired box 4 |
| chr7_-_125995884 | 0.69 |
ENSMUST00000075671.5
|
Nfatc2ip
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 interacting protein |
| chr12_-_32000534 | 0.66 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chrM_+_8603 | 0.62 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr9_-_123691077 | 0.51 |
ENSMUST00000182350.3
|
Xcr1
|
chemokine (C motif) receptor 1 |
| chr2_-_6327884 | 0.49 |
ENSMUST00000238876.2
|
1700014B07Rik
|
RIKEN cDNA 1700014B07 gene |
| chr10_+_80085275 | 0.49 |
ENSMUST00000020361.7
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr1_+_175459735 | 0.46 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr13_-_59930059 | 0.38 |
ENSMUST00000225581.2
|
Gm49354
|
predicted gene, 49354 |
| chr2_-_77349909 | 0.36 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr5_+_21391282 | 0.36 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr4_+_108317197 | 0.33 |
ENSMUST00000097925.9
|
Tut4
|
terminal uridylyl transferase 4 |
| chr1_+_52158721 | 0.29 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr2_-_77000878 | 0.16 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr7_+_130294403 | 0.07 |
ENSMUST00000207282.2
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr16_-_91443794 | 0.05 |
ENSMUST00000232367.2
ENSMUST00000231380.2 ENSMUST00000231444.2 ENSMUST00000232289.2 ENSMUST00000120450.2 ENSMUST00000023684.14 |
Gart
|
phosphoribosylglycinamide formyltransferase |
| chr7_-_80037688 | 0.00 |
ENSMUST00000080932.8
|
Fes
|
feline sarcoma oncogene |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.5 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 2.3 | 11.7 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 2.3 | 9.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.2 | 8.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 2.0 | 36.9 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 1.8 | 5.3 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 1.6 | 26.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.1 | 6.8 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 1.1 | 3.2 | GO:1990918 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.9 | 2.8 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.8 | 2.5 | GO:0061360 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.8 | 3.9 | GO:0009597 | detection of virus(GO:0009597) |
| 0.7 | 15.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.6 | 8.3 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.5 | 12.6 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.5 | 11.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 5.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) regulation of hematopoietic stem cell proliferation(GO:1902033) regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.4 | 2.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.4 | 1.6 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.4 | 6.9 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.3 | 1.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.3 | 7.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 | 4.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.3 | 1.5 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.3 | 6.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.3 | 6.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.3 | 4.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.3 | 1.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.3 | 3.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 0.9 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.2 | 3.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 3.8 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.2 | 4.8 | GO:0045056 | transcytosis(GO:0045056) |
| 0.2 | 13.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.1 | GO:0090160 | endosome localization(GO:0032439) Golgi to lysosome transport(GO:0090160) |
| 0.2 | 2.1 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.2 | 1.1 | GO:0035981 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.2 | 0.9 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 3.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 19.7 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.2 | 3.3 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 1.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 1.8 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 1.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 2.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 2.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.4 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.1 | 1.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 16.8 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 3.6 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 1.8 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 4.0 | GO:0045216 | cell-cell junction organization(GO:0045216) |
| 0.0 | 10.5 | GO:0006518 | peptide metabolic process(GO:0006518) |
| 0.0 | 0.8 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.5 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 1.8 | 60.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 1.4 | 4.3 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.5 | 6.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 16.9 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.2 | 10.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 2.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 40.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 44.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 3.6 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 1.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 16.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 4.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 11.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 10.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 10.2 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.6 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 11.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 3.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.3 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 1.3 | 11.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.3 | 5.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.9 | 8.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.9 | 10.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.8 | 25.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.7 | 12.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.5 | 13.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.5 | 13.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.5 | 9.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 11.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 2.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.3 | 3.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 3.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 4.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 2.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 9.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 1.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 2.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 1.6 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.2 | 1.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 4.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 2.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 26.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 3.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 3.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 51.8 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 6.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 12.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 6.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 2.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 36.9 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 3.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 26.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 36.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.3 | 2.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 22.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 9.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 4.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 6.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 11.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 5.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 2.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 8.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 10.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 26.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.8 | 40.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.1 | 23.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.5 | 19.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 11.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 4.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 9.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 3.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 2.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 4.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 2.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 5.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 2.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |