Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Irx4
Z-value: 0.51
Transcription factors associated with Irx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irx4
|
ENSMUSG00000021604.16 | Irx4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx4 | mm39_v1_chr13_+_73408591_73408616 | 0.10 | 4.3e-01 | Click! |
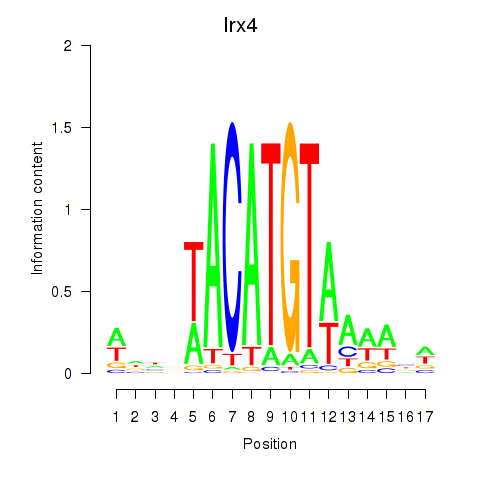
Activity profile of Irx4 motif
Sorted Z-values of Irx4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_105048856 | 4.40 |
ENSMUST00000041973.7
|
Cmtm2b
|
CKLF-like MARVEL transmembrane domain containing 2B |
| chr6_-_3494587 | 4.27 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr8_-_25215778 | 3.71 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr18_+_44467133 | 3.69 |
ENSMUST00000025349.12
ENSMUST00000115498.2 |
Myot
|
myotilin |
| chr4_+_48663504 | 3.27 |
ENSMUST00000030033.5
|
Cavin4
|
caveolae associated 4 |
| chr8_-_25215856 | 3.07 |
ENSMUST00000033958.15
|
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr1_+_45350698 | 3.07 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr11_-_11920540 | 2.73 |
ENSMUST00000109653.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr16_+_16964801 | 1.99 |
ENSMUST00000232479.2
ENSMUST00000232344.2 ENSMUST00000069064.7 |
Ydjc
|
YdjC homolog (bacterial) |
| chr12_-_114252202 | 1.84 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr6_-_69584812 | 1.49 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr2_-_79738773 | 1.37 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr8_+_27937128 | 1.24 |
ENSMUST00000209669.2
|
Gm45861
|
predicted gene 45861 |
| chr3_+_27371206 | 1.19 |
ENSMUST00000174840.2
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr2_-_79738734 | 1.07 |
ENSMUST00000090756.11
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_+_24235300 | 1.02 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr7_+_106413336 | 0.95 |
ENSMUST00000166880.3
ENSMUST00000075414.8 |
Olfr701
|
olfactory receptor 701 |
| chr3_+_27371168 | 0.90 |
ENSMUST00000046383.12
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr15_-_35938155 | 0.90 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr17_-_27352876 | 0.83 |
ENSMUST00000119227.3
ENSMUST00000025045.15 |
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr15_-_35938328 | 0.82 |
ENSMUST00000014457.15
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chrY_-_797405 | 0.75 |
ENSMUST00000189888.7
ENSMUST00000065545.6 |
Zfy1
|
zinc finger protein 1, Y-linked |
| chr6_-_83294526 | 0.72 |
ENSMUST00000005810.9
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr3_+_104974160 | 0.53 |
ENSMUST00000070584.5
|
4930564D02Rik
|
RIKEN cDNA 4930564D02 gene |
| chr9_-_58108988 | 0.38 |
ENSMUST00000163200.3
ENSMUST00000165276.2 ENSMUST00000214647.2 |
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr9_-_58109564 | 0.36 |
ENSMUST00000163897.8
ENSMUST00000215950.2 |
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr3_+_59832635 | 0.34 |
ENSMUST00000049476.3
|
Aadacl2fm1
|
AADACL2 family member 1 |
| chr3_+_59637203 | 0.19 |
ENSMUST00000168156.3
|
Aadacl2fm2
|
AADACL2 family member 2 |
| chr2_-_111253457 | 0.19 |
ENSMUST00000213210.2
ENSMUST00000184954.4 |
Olfr1286
|
olfactory receptor 1286 |
| chr12_-_115825934 | 0.18 |
ENSMUST00000198777.2
|
Ighv1-77
|
immunoglobulin heavy variable 1-77 |
| chr3_+_84832783 | 0.15 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr9_+_19716202 | 0.08 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr12_-_111946560 | 0.05 |
ENSMUST00000190680.2
|
Rd3l
|
retinal degeneration 3-like |
| chr8_+_105048199 | 0.05 |
ENSMUST00000109447.2
|
Gm11020
|
predicted gene 11020 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 2.7 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 1.0 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 6.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 3.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 2.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.8 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 2.1 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 4.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.7 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 2.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 7.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 4.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 6.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.2 | 2.4 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 3.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.7 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 1.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 3.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 3.5 | GO:0005125 | cytokine activity(GO:0005125) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 3.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |