Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
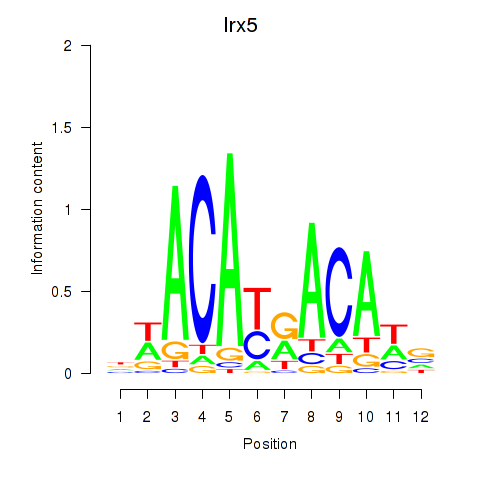
Results for Irx5
Z-value: 1.12
Transcription factors associated with Irx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irx5
|
ENSMUSG00000031737.12 | Irx5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx5 | mm39_v1_chr8_+_93084253_93084379 | 0.12 | 3.2e-01 | Click! |
Activity profile of Irx5 motif
Sorted Z-values of Irx5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_87074380 | 12.46 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chrX_+_20416019 | 10.47 |
ENSMUST00000023832.7
|
Rgn
|
regucalcin |
| chr15_+_82336535 | 10.13 |
ENSMUST00000089129.7
ENSMUST00000229313.2 ENSMUST00000231136.2 |
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr4_+_104623505 | 9.79 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr7_-_14172434 | 9.12 |
ENSMUST00000210396.2
ENSMUST00000168252.9 |
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr8_-_45715049 | 8.54 |
ENSMUST00000034064.5
|
F11
|
coagulation factor XI |
| chr5_-_87485023 | 7.38 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr5_-_87402659 | 7.37 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr3_+_20011201 | 6.99 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr19_-_7779943 | 6.77 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr15_-_82648376 | 6.60 |
ENSMUST00000055721.6
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr11_-_110142565 | 6.54 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr4_+_141473983 | 5.66 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr8_-_73188887 | 5.46 |
ENSMUST00000109974.2
|
Calr3
|
calreticulin 3 |
| chr3_+_20011405 | 5.06 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr13_+_25127127 | 4.74 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr3_+_20011251 | 4.62 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr2_-_101451383 | 4.19 |
ENSMUST00000090513.11
|
Iftap
|
intraflagellar transport associated protein |
| chr2_+_172994841 | 3.97 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr15_-_82278223 | 3.85 |
ENSMUST00000170255.2
|
Cyp2d11
|
cytochrome P450, family 2, subfamily d, polypeptide 11 |
| chr8_-_58249608 | 3.84 |
ENSMUST00000204067.3
|
Galntl6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr14_-_59380335 | 3.78 |
ENSMUST00000022548.10
ENSMUST00000162674.8 ENSMUST00000159858.2 ENSMUST00000162271.2 |
1700129C05Rik
|
RIKEN cDNA 1700129C05 gene |
| chr19_-_28945194 | 3.47 |
ENSMUST00000162110.8
|
Spata6l
|
spermatogenesis associated 6 like |
| chr17_+_72076678 | 3.47 |
ENSMUST00000230427.2
ENSMUST00000229952.2 ENSMUST00000230333.2 |
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr6_+_50087826 | 3.40 |
ENSMUST00000167628.2
|
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr17_+_72076728 | 3.28 |
ENSMUST00000230305.2
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr11_-_58346806 | 3.10 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr1_-_160986880 | 3.06 |
ENSMUST00000135643.8
ENSMUST00000178511.3 |
Tex50
|
testis expressed 50 |
| chr2_-_73456080 | 3.05 |
ENSMUST00000070579.7
|
Chn1
|
chimerin 1 |
| chr11_-_103395423 | 2.95 |
ENSMUST00000153273.3
|
Lrrc37a
|
leucine rich repeat containing 37A |
| chr2_+_116709167 | 2.92 |
ENSMUST00000123598.8
ENSMUST00000155470.8 |
Tmco5
|
transmembrane and coiled-coil domains 5 |
| chr18_-_35348049 | 2.79 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr19_+_39499288 | 2.76 |
ENSMUST00000025968.5
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr10_+_86614864 | 2.69 |
ENSMUST00000099396.3
|
Nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr3_+_138911648 | 2.67 |
ENSMUST00000062306.7
|
Stpg2
|
sperm tail PG rich repeat containing 2 |
| chr4_-_58009118 | 2.56 |
ENSMUST00000102897.11
ENSMUST00000239406.2 |
Txndc8
|
thioredoxin domain containing 8 |
| chr6_-_129303659 | 2.47 |
ENSMUST00000203159.2
|
Clec2m
|
C-type lectin domain family 2, member m |
| chr14_+_48358267 | 2.46 |
ENSMUST00000073150.6
|
Peli2
|
pellino 2 |
| chr3_-_126792056 | 2.45 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr11_+_78067537 | 2.42 |
ENSMUST00000098545.12
|
Tlcd1
|
TLC domain containing 1 |
| chr1_+_132155922 | 2.40 |
ENSMUST00000191418.2
|
Lemd1
|
LEM domain containing 1 |
| chr14_-_4506874 | 2.34 |
ENSMUST00000224934.2
|
Thrb
|
thyroid hormone receptor beta |
| chr19_+_26600820 | 2.30 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_+_107655487 | 2.29 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr3_+_87265181 | 2.29 |
ENSMUST00000015998.8
|
Cd5l
|
CD5 antigen-like |
| chr3_-_88669551 | 2.14 |
ENSMUST00000183267.2
|
Syt11
|
synaptotagmin XI |
| chr6_-_113694633 | 2.12 |
ENSMUST00000204533.3
|
Ghrl
|
ghrelin |
| chr11_+_42312150 | 2.10 |
ENSMUST00000192403.2
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr11_-_69286159 | 2.01 |
ENSMUST00000108660.8
ENSMUST00000051620.5 |
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr10_-_63039709 | 2.00 |
ENSMUST00000095580.3
|
Mypn
|
myopalladin |
| chr12_+_85335365 | 1.95 |
ENSMUST00000059341.5
|
Zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr12_-_85335193 | 1.85 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr4_-_43653539 | 1.82 |
ENSMUST00000084646.11
|
Spag8
|
sperm associated antigen 8 |
| chr2_-_111062182 | 1.79 |
ENSMUST00000099620.5
|
Olfr1275
|
olfactory receptor 1275 |
| chr12_-_67269323 | 1.79 |
ENSMUST00000037181.16
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr5_-_121641461 | 1.79 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr8_-_43594523 | 1.78 |
ENSMUST00000059692.4
|
Triml1
|
tripartite motif family-like 1 |
| chr11_-_121410152 | 1.77 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chr1_+_86354045 | 1.66 |
ENSMUST00000046004.6
|
Tex44
|
testis expressed 44 |
| chr2_+_121991181 | 1.63 |
ENSMUST00000036089.8
|
Trim69
|
tripartite motif-containing 69 |
| chr14_-_70449438 | 1.63 |
ENSMUST00000227929.2
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr2_+_152269532 | 1.63 |
ENSMUST00000128737.2
|
6820408C15Rik
|
RIKEN cDNA 6820408C15 gene |
| chr4_-_43653559 | 1.63 |
ENSMUST00000107870.3
|
Spag8
|
sperm associated antigen 8 |
| chr4_-_114991174 | 1.58 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr1_-_169796709 | 1.57 |
ENSMUST00000027989.13
ENSMUST00000111353.4 |
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr11_+_74174562 | 1.50 |
ENSMUST00000214048.3
ENSMUST00000143976.4 ENSMUST00000205790.2 ENSMUST00000206659.2 |
Olfr59
|
olfactory receptor 59 |
| chr11_+_69286473 | 1.50 |
ENSMUST00000144531.2
|
Naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chrX_+_133088238 | 1.46 |
ENSMUST00000064476.5
|
Arl13a
|
ADP-ribosylation factor-like 13A |
| chr7_-_86016045 | 1.43 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr2_-_136229849 | 1.42 |
ENSMUST00000035264.9
ENSMUST00000077200.4 |
Pak5
|
p21 (RAC1) activated kinase 5 |
| chr11_+_68989763 | 1.33 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr5_-_74189898 | 1.31 |
ENSMUST00000152408.3
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr4_+_109092459 | 1.31 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chrX_+_37861548 | 1.21 |
ENSMUST00000050744.6
|
6030498E09Rik
|
RIKEN cDNA 6030498E09 gene |
| chr9_+_110948492 | 1.20 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr19_+_6130059 | 1.17 |
ENSMUST00000149347.8
ENSMUST00000143303.2 |
Tmem262
|
transmembrane protein 262 |
| chr11_+_62737887 | 1.12 |
ENSMUST00000036085.11
|
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr7_+_55889488 | 1.10 |
ENSMUST00000032633.12
ENSMUST00000155533.2 ENSMUST00000156886.8 |
Oca2
|
oculocutaneous albinism II |
| chr7_+_102289455 | 1.07 |
ENSMUST00000098221.2
|
Olfr554
|
olfactory receptor 554 |
| chr13_+_22462487 | 1.06 |
ENSMUST00000091736.5
|
Vmn1r195
|
vomeronasal 1 receptor 195 |
| chr16_+_3408906 | 1.01 |
ENSMUST00000216259.2
|
Olfr161
|
olfactory receptor 161 |
| chr10_-_99595498 | 1.00 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr2_+_85409559 | 0.99 |
ENSMUST00000077075.5
|
Olfr996
|
olfactory receptor 996 |
| chr11_+_62737936 | 0.99 |
ENSMUST00000150989.8
ENSMUST00000176577.2 |
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr11_+_73244561 | 0.96 |
ENSMUST00000108465.4
|
Olfr20
|
olfactory receptor 20 |
| chr2_+_89711138 | 0.90 |
ENSMUST00000060795.3
|
Olfr1257
|
olfactory receptor 1257 |
| chr2_+_103858066 | 0.89 |
ENSMUST00000028603.10
|
Fbxo3
|
F-box protein 3 |
| chr7_+_102319108 | 0.89 |
ENSMUST00000098219.3
|
Olfr556
|
olfactory receptor 556 |
| chr3_-_32419609 | 0.87 |
ENSMUST00000139660.2
ENSMUST00000168566.3 ENSMUST00000029199.11 |
Zmat3
|
zinc finger matrin type 3 |
| chr4_+_8535604 | 0.81 |
ENSMUST00000060232.8
|
Rab2a
|
RAB2A, member RAS oncogene family |
| chr10_+_129376408 | 0.80 |
ENSMUST00000076575.4
|
Olfr792
|
olfactory receptor 792 |
| chr6_+_71470987 | 0.80 |
ENSMUST00000114179.3
|
Rnf103
|
ring finger protein 103 |
| chr7_-_106341650 | 0.79 |
ENSMUST00000217734.2
|
Olfr697
|
olfactory receptor 697 |
| chr8_+_4290105 | 0.76 |
ENSMUST00000110994.9
ENSMUST00000110995.8 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr2_-_85478663 | 0.71 |
ENSMUST00000099920.2
|
Olfr1002
|
olfactory receptor 1002 |
| chr7_+_48038274 | 0.67 |
ENSMUST00000056676.5
|
Mrgprb8
|
MAS-related GPR, member B8 |
| chr12_-_108859123 | 0.66 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr12_-_81426238 | 0.64 |
ENSMUST00000062182.8
|
Gm4787
|
predicted gene 4787 |
| chr7_-_10292412 | 0.62 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr14_-_40726472 | 0.58 |
ENSMUST00000153830.8
|
Prxl2a
|
peroxiredoxin like 2A |
| chr10_+_129611696 | 0.56 |
ENSMUST00000059891.3
ENSMUST00000218237.2 |
Olfr809
|
olfactory receptor 809 |
| chr6_+_40470463 | 0.52 |
ENSMUST00000038750.7
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr9_-_18667005 | 0.51 |
ENSMUST00000066997.4
|
Olfr24
|
olfactory receptor 24 |
| chr2_+_85597442 | 0.48 |
ENSMUST00000216397.3
|
Olfr1013
|
olfactory receptor 1013 |
| chr9_+_37683153 | 0.47 |
ENSMUST00000215128.2
|
Olfr875
|
olfactory receptor 875 |
| chr16_-_22630327 | 0.46 |
ENSMUST00000040592.6
|
Crygs
|
crystallin, gamma S |
| chr14_+_8348779 | 0.43 |
ENSMUST00000022256.5
|
Psmd6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr6_-_57938488 | 0.42 |
ENSMUST00000228097.2
|
Vmn1r24
|
vomeronasal 1 receptor 24 |
| chr12_+_29988035 | 0.41 |
ENSMUST00000122328.8
ENSMUST00000118321.3 |
Pxdn
|
peroxidasin |
| chr2_+_103858168 | 0.40 |
ENSMUST00000111135.8
ENSMUST00000111136.8 ENSMUST00000102565.4 |
Fbxo3
|
F-box protein 3 |
| chr6_+_56926317 | 0.40 |
ENSMUST00000227073.2
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr11_+_74150634 | 0.39 |
ENSMUST00000217016.2
ENSMUST00000214303.2 |
Olfr406
|
olfactory receptor 406 |
| chr17_-_35954573 | 0.38 |
ENSMUST00000095467.4
|
Mucl3
|
mucin like 3 |
| chr7_-_5325456 | 0.37 |
ENSMUST00000207520.2
|
Nlrp2
|
NLR family, pyrin domain containing 2 |
| chr7_-_66915756 | 0.34 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr19_-_55229668 | 0.34 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr4_-_43700807 | 0.29 |
ENSMUST00000055545.5
|
Olfr70
|
olfactory receptor 70 |
| chr19_+_12186932 | 0.29 |
ENSMUST00000072316.5
|
Olfr1431
|
olfactory receptor 1431 |
| chr10_+_97400990 | 0.28 |
ENSMUST00000038160.6
|
Lum
|
lumican |
| chr19_+_13745671 | 0.28 |
ENSMUST00000061669.3
|
Olfr1495
|
olfactory receptor 1495 |
| chr15_-_76906832 | 0.28 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr7_-_133384449 | 0.26 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr17_+_87332978 | 0.25 |
ENSMUST00000024959.10
|
Cript
|
cysteine-rich PDZ-binding protein |
| chr19_-_12219023 | 0.23 |
ENSMUST00000087818.2
|
Olfr262
|
olfactory receptor 262 |
| chr6_+_56974228 | 0.20 |
ENSMUST00000228285.2
ENSMUST00000227847.2 ENSMUST00000226689.2 ENSMUST00000227131.2 ENSMUST00000227631.2 ENSMUST00000227188.2 |
Vmn1r6
|
vomeronasal 1 receptor 6 |
| chr10_+_3822667 | 0.17 |
ENSMUST00000136671.8
ENSMUST00000042438.13 |
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr17_-_27841759 | 0.14 |
ENSMUST00000176458.2
ENSMUST00000114886.8 |
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr4_+_148123554 | 0.12 |
ENSMUST00000141283.8
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr2_-_155661324 | 0.12 |
ENSMUST00000124586.2
|
BC029722
|
cDNA sequence BC029722 |
| chr6_-_57306479 | 0.06 |
ENSMUST00000227283.2
ENSMUST00000228356.2 |
Vmn1r16
|
vomeronasal 1 receptor 16 |
| chr3_-_79645101 | 0.03 |
ENSMUST00000078527.13
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr17_-_87332780 | 0.03 |
ENSMUST00000024957.7
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr18_+_44403169 | 0.02 |
ENSMUST00000042747.4
|
Npy6r
|
neuropeptide Y receptor Y6 |
| chr17_+_33395390 | 0.01 |
ENSMUST00000112168.4
|
Olfr55
|
olfactory receptor 55 |
| chr18_+_37939442 | 0.01 |
ENSMUST00000076807.7
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 2.6 | 10.5 | GO:1903630 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 1.7 | 8.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.2 | 4.7 | GO:0010982 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 1.0 | 4.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.9 | 5.7 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.6 | 9.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 2.1 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.5 | 2.1 | GO:1904344 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 0.5 | 7.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.5 | 6.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.5 | 16.7 | GO:0046688 | copper ion transport(GO:0006825) response to copper ion(GO:0046688) |
| 0.4 | 2.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.3 | 2.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 2.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 19.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 1.2 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.2 | 2.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 1.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 0.7 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 2.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 1.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 2.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 2.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 3.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.8 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.1 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 1.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 5.5 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 2.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 3.3 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 1.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 1.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 5.6 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.0 | GO:0060618 | nipple development(GO:0060618) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 1.5 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 16.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 4.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 2.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 4.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 7.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 16.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.3 | 4.0 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 2.1 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.7 | 27.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.5 | 4.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.5 | 1.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.5 | 18.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.5 | 5.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 8.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.5 | 1.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.4 | 6.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 2.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 2.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 2.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 0.7 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 5.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 3.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.8 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.4 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 2.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 2.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 2.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 6.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 6.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 2.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 3.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 16.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 3.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.4 | 8.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 9.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 5.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 6.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 4.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 2.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |