Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Irx6_Irx2_Irx3
Z-value: 1.07
Transcription factors associated with Irx6_Irx2_Irx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
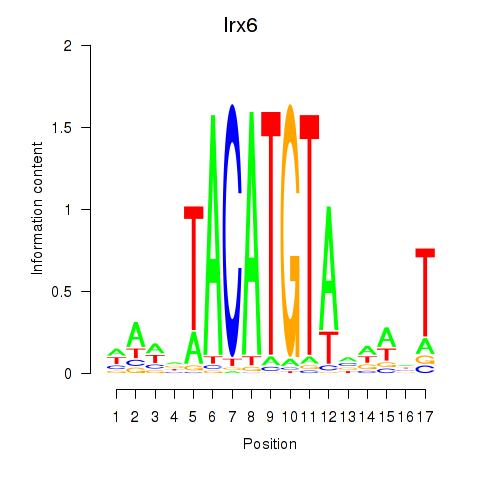
Irx6
|
ENSMUSG00000031738.15 | Irx6 |
|
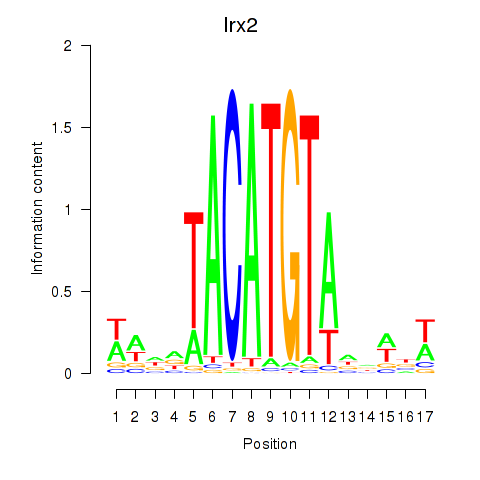
Irx2
|
ENSMUSG00000001504.11 | Irx2 |
|
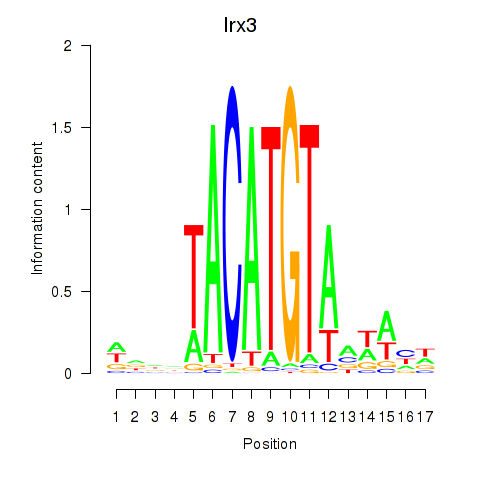
Irx3
|
ENSMUSG00000031734.14 | Irx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx2 | mm39_v1_chr13_+_72776921_72776967 | 0.32 | 5.9e-03 | Click! |
| Irx6 | mm39_v1_chr8_+_93401454_93401454 | -0.28 | 1.6e-02 | Click! |
| Irx3 | mm39_v1_chr8_-_92528570_92528695 | 0.17 | 1.5e-01 | Click! |
Activity profile of Irx6_Irx2_Irx3 motif
Sorted Z-values of Irx6_Irx2_Irx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx6_Irx2_Irx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_44096300 | 8.53 |
ENSMUST00000069245.8
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr6_-_129449739 | 6.96 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr8_+_105048856 | 6.60 |
ENSMUST00000041973.7
|
Cmtm2b
|
CKLF-like MARVEL transmembrane domain containing 2B |
| chr6_+_42015319 | 5.22 |
ENSMUST00000117406.8
ENSMUST00000024059.5 |
Sva
|
seminal vesicle antigen |
| chr5_-_87847268 | 4.03 |
ENSMUST00000196869.5
ENSMUST00000199624.5 ENSMUST00000198057.5 ENSMUST00000082370.10 |
Csn2
|
casein beta |
| chr11_-_97886997 | 4.02 |
ENSMUST00000042971.16
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr6_-_48685108 | 3.90 |
ENSMUST00000126422.3
ENSMUST00000119315.2 ENSMUST00000053661.7 |
Gimap6
|
GTPase, IMAP family member 6 |
| chrX_+_106132055 | 3.79 |
ENSMUST00000150494.2
|
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr4_+_111863441 | 3.58 |
ENSMUST00000162885.8
ENSMUST00000117379.9 ENSMUST00000161389.8 ENSMUST00000162158.2 |
Skint1
|
selection and upkeep of intraepithelial T cells 1 |
| chr15_-_76906832 | 3.11 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr4_+_97665843 | 3.05 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr3_+_59914164 | 2.97 |
ENSMUST00000169794.2
|
Aadacl2
|
arylacetamide deacetylase like 2 |
| chr3_-_122828592 | 2.81 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr1_+_152626273 | 2.63 |
ENSMUST00000068875.5
|
Apobec4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr8_+_85635189 | 2.53 |
ENSMUST00000003910.13
ENSMUST00000109744.8 |
Dnase2a
|
deoxyribonuclease II alpha |
| chr11_+_14549252 | 2.43 |
ENSMUST00000117584.3
|
Pom121l12
|
POM121 membrane glycoprotein-like 12 |
| chr6_-_130363837 | 2.39 |
ENSMUST00000032288.6
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr10_+_61007733 | 2.36 |
ENSMUST00000122261.8
ENSMUST00000121297.8 ENSMUST00000035894.12 |
Tbata
|
thymus, brain and testes associated |
| chr16_+_49620883 | 2.26 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr7_-_24033346 | 2.17 |
ENSMUST00000069562.6
|
Tescl
|
tescalcin-like |
| chr3_-_92627651 | 2.17 |
ENSMUST00000047153.4
|
Lce1f
|
late cornified envelope 1F |
| chr8_-_105019806 | 2.16 |
ENSMUST00000212492.2
ENSMUST00000034344.10 |
Cmtm2a
|
CKLF-like MARVEL transmembrane domain containing 2A |
| chr6_+_41039255 | 2.15 |
ENSMUST00000103266.3
|
Trbv5
|
T cell receptor beta, variable 5 |
| chr17_+_34364206 | 2.12 |
ENSMUST00000041982.9
ENSMUST00000171231.8 |
H2-DMb2
|
histocompatibility 2, class II, locus Mb2 |
| chr17_+_30845909 | 2.08 |
ENSMUST00000236140.2
ENSMUST00000236118.2 ENSMUST00000235390.2 |
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr14_+_53859114 | 2.07 |
ENSMUST00000103657.6
|
Trav12-3
|
T cell receptor alpha variable 12-3 |
| chr9_-_36549886 | 2.07 |
ENSMUST00000093868.9
ENSMUST00000184611.2 |
Pate14
|
prostate and testis expressed 14 |
| chr9_-_44473146 | 2.05 |
ENSMUST00000215293.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr6_+_41204430 | 2.04 |
ENSMUST00000193064.2
ENSMUST00000103280.3 |
Trbv26
|
T cell receptor beta, variable 26 |
| chr14_+_53257873 | 2.02 |
ENSMUST00000196756.2
|
Trav7d-6
|
T cell receptor alpha variable 7D-6 |
| chr8_+_72050292 | 2.02 |
ENSMUST00000143662.8
|
Niban3
|
niban apoptosis regulator 3 |
| chr1_-_72739704 | 2.00 |
ENSMUST00000053499.6
ENSMUST00000212710.2 ENSMUST00000212573.2 |
Ankar
|
ankyrin and armadillo repeat containing |
| chr8_-_25215778 | 1.99 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chrX_+_94601705 | 1.99 |
ENSMUST00000210331.2
|
Pfn5
|
profilin 5 |
| chr7_-_103477126 | 1.98 |
ENSMUST00000023934.8
|
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr3_+_59832635 | 1.96 |
ENSMUST00000049476.3
|
Aadacl2fm1
|
AADACL2 family member 1 |
| chr1_-_173703424 | 1.91 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr6_-_130208601 | 1.91 |
ENSMUST00000088011.11
ENSMUST00000112013.8 ENSMUST00000049304.14 |
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr14_+_44340111 | 1.91 |
ENSMUST00000074839.7
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr2_+_24235300 | 1.91 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr8_-_25215856 | 1.91 |
ENSMUST00000033958.15
|
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr13_+_19398273 | 1.89 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chrX_+_54272236 | 1.87 |
ENSMUST00000088740.5
|
Slxl1
|
Slx-like 1 |
| chr14_+_53478202 | 1.86 |
ENSMUST00000179583.3
|
Trav12n-3
|
T cell receptor alpha variable 12N-3 |
| chr17_+_30843328 | 1.78 |
ENSMUST00000170651.2
|
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr14_+_53448363 | 1.72 |
ENSMUST00000103618.2
|
Trav4n-3
|
T cell receptor alpha variable 4N-3 |
| chr1_-_72739691 | 1.72 |
ENSMUST00000211837.2
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr10_+_102376109 | 1.71 |
ENSMUST00000055355.6
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr11_+_11414256 | 1.69 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr13_-_35100960 | 1.68 |
ENSMUST00000225242.2
|
Fam217a
|
family with sequence similarity 217, member A |
| chr15_+_79775819 | 1.67 |
ENSMUST00000177350.8
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr10_+_111919250 | 1.67 |
ENSMUST00000148897.8
ENSMUST00000020434.4 |
Glipr1l2
|
GLI pathogenesis-related 1 like 2 |
| chr14_+_53137426 | 1.67 |
ENSMUST00000103592.2
|
Trav4d-3
|
T cell receptor alpha variable 4D-3 |
| chr9_-_99592116 | 1.64 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr18_+_44467133 | 1.64 |
ENSMUST00000025349.12
ENSMUST00000115498.2 |
Myot
|
myotilin |
| chr14_+_53836282 | 1.63 |
ENSMUST00000103655.3
|
Trav4-3
|
T cell receptor alpha variable 4-3 |
| chr14_-_44161016 | 1.62 |
ENSMUST00000159175.2
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10 |
| chr12_-_101784727 | 1.61 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chrX_+_53533175 | 1.59 |
ENSMUST00000180150.8
|
Gm16405
|
predicted gene 16405 |
| chrX_+_53624178 | 1.59 |
ENSMUST00000178145.8
|
Gm16430
|
predicted gene 16430 |
| chr1_-_159981132 | 1.57 |
ENSMUST00000039178.12
|
Tnn
|
tenascin N |
| chr10_-_125164826 | 1.57 |
ENSMUST00000211781.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr16_-_64422716 | 1.57 |
ENSMUST00000209382.3
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr5_-_18054702 | 1.55 |
ENSMUST00000165232.8
|
Cd36
|
CD36 molecule |
| chr8_+_85598734 | 1.53 |
ENSMUST00000170296.2
ENSMUST00000136026.8 |
Syce2
|
synaptonemal complex central element protein 2 |
| chr5_-_18054781 | 1.51 |
ENSMUST00000170051.8
|
Cd36
|
CD36 molecule |
| chr6_-_41535322 | 1.51 |
ENSMUST00000193003.2
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr6_+_41036271 | 1.50 |
ENSMUST00000103265.5
|
Trbv4
|
T cell receptor beta, variable 10 |
| chr14_+_54068562 | 1.45 |
ENSMUST00000103673.11
ENSMUST00000186573.2 |
Trav18
|
T cell receptor alpha variable 18 |
| chr13_+_19528728 | 1.45 |
ENSMUST00000179181.3
|
Trgc4
|
T cell receptor gamma, constant 4 |
| chr6_-_41535292 | 1.44 |
ENSMUST00000103300.3
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr14_-_44057096 | 1.44 |
ENSMUST00000100691.4
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr1_-_130557349 | 1.41 |
ENSMUST00000142416.2
ENSMUST00000039862.11 ENSMUST00000128128.8 |
Zp3r
|
zona pellucida 3 receptor |
| chr1_+_131566044 | 1.38 |
ENSMUST00000073350.13
|
Ctse
|
cathepsin E |
| chr3_+_106020545 | 1.37 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr2_-_140231618 | 1.36 |
ENSMUST00000122367.8
ENSMUST00000120133.2 |
Sel1l2
|
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr3_+_84573499 | 1.36 |
ENSMUST00000107682.2
|
Tmem154
|
transmembrane protein 154 |
| chr1_-_28819331 | 1.32 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr5_-_86893645 | 1.31 |
ENSMUST00000161306.2
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr12_+_119407145 | 1.29 |
ENSMUST00000048880.7
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr8_-_86091970 | 1.28 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_+_85635341 | 1.27 |
ENSMUST00000134569.2
|
Dnase2a
|
deoxyribonuclease II alpha |
| chr8_+_13087292 | 1.27 |
ENSMUST00000063820.12
ENSMUST00000033821.11 |
F10
|
coagulation factor X |
| chr6_-_130044234 | 1.26 |
ENSMUST00000119096.2
|
Klra4
|
killer cell lectin-like receptor, subfamily A, member 4 |
| chr2_+_154032731 | 1.26 |
ENSMUST00000081816.11
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr6_-_70895899 | 1.25 |
ENSMUST00000063456.5
|
Tex37
|
testis expressed 37 |
| chr7_+_44926925 | 1.25 |
ENSMUST00000210861.2
|
Slc6a21
|
solute carrier family 6 member 21 |
| chr6_-_128700022 | 1.23 |
ENSMUST00000112110.4
|
Klrb1
|
killer cell lectin-like receptor subfamily B member 1 |
| chr4_-_43483696 | 1.23 |
ENSMUST00000030180.7
|
Sit1
|
suppression inducing transmembrane adaptor 1 |
| chr1_-_128520002 | 1.21 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr1_+_45350698 | 1.20 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr8_+_81220410 | 1.20 |
ENSMUST00000063359.8
|
Gypa
|
glycophorin A |
| chr6_-_56878854 | 1.16 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr6_-_23655130 | 1.16 |
ENSMUST00000104979.2
|
Rnf148
|
ring finger protein 148 |
| chr6_+_41248311 | 1.15 |
ENSMUST00000103281.3
|
Trbv29
|
T cell receptor beta, variable 29 |
| chr13_-_21937997 | 1.15 |
ENSMUST00000074752.4
|
H2ac15
|
H2A clustered histone 15 |
| chr13_-_24945844 | 1.14 |
ENSMUST00000006898.10
ENSMUST00000110382.9 |
Gmnn
|
geminin |
| chr2_-_119101680 | 1.13 |
ENSMUST00000037360.8
|
Rhov
|
ras homolog family member V |
| chr6_-_113696390 | 1.13 |
ENSMUST00000203588.2
ENSMUST00000204163.3 ENSMUST00000203363.3 |
Ghrl
|
ghrelin |
| chr9_-_35481689 | 1.13 |
ENSMUST00000115110.5
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr11_+_88964667 | 1.12 |
ENSMUST00000100619.11
|
Gm525
|
predicted gene 525 |
| chr13_-_19495474 | 1.11 |
ENSMUST00000197113.2
|
Trgj2
|
T cell receptor gamma joining 2 |
| chr7_-_3551003 | 1.11 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr8_-_79539838 | 1.11 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr13_+_19394484 | 1.10 |
ENSMUST00000200495.2
|
Trgj1
|
T cell receptor gamma joining 1 |
| chr6_+_122929627 | 1.08 |
ENSMUST00000204427.2
|
Clec4a3
|
C-type lectin domain family 4, member a3 |
| chrX_-_100307592 | 1.08 |
ENSMUST00000101358.3
|
Gm614
|
predicted gene 614 |
| chr10_-_23663109 | 1.08 |
ENSMUST00000218221.2
ENSMUST00000218107.2 |
Rps12
|
ribosomal protein S12 |
| chr6_-_116561156 | 1.08 |
ENSMUST00000061723.6
|
Olfr215
|
olfactory receptor 215 |
| chrX_+_143253677 | 1.06 |
ENSMUST00000178233.2
|
Trpc5os
|
transient receptor potential cation channel, subfamily C, member 5, opposite strand |
| chrY_-_797405 | 1.04 |
ENSMUST00000189888.7
ENSMUST00000065545.6 |
Zfy1
|
zinc finger protein 1, Y-linked |
| chr14_+_53167246 | 1.03 |
ENSMUST00000177703.3
|
Trav12d-3
|
T cell receptor alpha variable 12D-3 |
| chr10_+_26105605 | 1.03 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr3_+_92840279 | 1.02 |
ENSMUST00000046234.5
|
Lce3b
|
late cornified envelope 3B |
| chr15_+_91083678 | 1.02 |
ENSMUST00000190436.7
|
CN725425
|
cDNA sequence CN725425 |
| chr16_+_35977170 | 1.02 |
ENSMUST00000079184.6
|
Stfa2l1
|
stefin A2 like 1 |
| chr19_-_10655391 | 1.01 |
ENSMUST00000025647.7
|
Pga5
|
pepsinogen 5, group I |
| chr7_+_106413336 | 1.01 |
ENSMUST00000166880.3
ENSMUST00000075414.8 |
Olfr701
|
olfactory receptor 701 |
| chr7_-_130148984 | 1.01 |
ENSMUST00000160289.9
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr5_+_77184892 | 0.99 |
ENSMUST00000118941.2
|
Thegl
|
theg spermatid protein like |
| chr3_+_92259448 | 0.99 |
ENSMUST00000068399.2
|
Sprr2e
|
small proline-rich protein 2E |
| chr7_-_119078472 | 0.98 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr5_+_123214332 | 0.98 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr14_-_44112974 | 0.96 |
ENSMUST00000179200.2
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr14_-_68893253 | 0.96 |
ENSMUST00000225767.3
ENSMUST00000111072.8 ENSMUST00000022642.6 ENSMUST00000224039.2 |
Adam28
|
a disintegrin and metallopeptidase domain 28 |
| chr10_+_63265871 | 0.95 |
ENSMUST00000105441.8
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr6_+_30512285 | 0.95 |
ENSMUST00000031798.14
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr10_-_38998272 | 0.95 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr16_-_19801781 | 0.94 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chrX_-_56326951 | 0.94 |
ENSMUST00000114735.9
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr14_+_53562089 | 0.94 |
ENSMUST00000178100.3
|
Trav7n-6
|
T cell receptor alpha variable 7N-6 |
| chr6_-_83294526 | 0.94 |
ENSMUST00000005810.9
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr1_-_44140820 | 0.92 |
ENSMUST00000152239.8
|
Tex30
|
testis expressed 30 |
| chr1_+_190769010 | 0.92 |
ENSMUST00000077889.8
|
Spata45
|
spermatogenesis associated 45 |
| chr17_+_41121979 | 0.92 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr12_-_114728503 | 0.92 |
ENSMUST00000194625.2
|
Ighv1-23
|
immunoglobulin heavy variable V1-23 |
| chr13_-_26954110 | 0.92 |
ENSMUST00000055915.6
|
Hdgfl1
|
HDGF like 1 |
| chr2_-_17395765 | 0.91 |
ENSMUST00000177966.2
|
Nebl
|
nebulette |
| chr11_-_106205320 | 0.91 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr6_-_113696809 | 0.91 |
ENSMUST00000203770.3
ENSMUST00000064993.8 |
Ghrl
|
ghrelin |
| chr5_-_77359039 | 0.90 |
ENSMUST00000121825.2
|
Spink2
|
serine peptidase inhibitor, Kazal type 2 |
| chr1_-_138103021 | 0.90 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr8_+_85635311 | 0.90 |
ENSMUST00000145292.8
|
Dnase2a
|
deoxyribonuclease II alpha |
| chr1_-_174078542 | 0.90 |
ENSMUST00000061990.5
|
Olfr419
|
olfactory receptor 419 |
| chr6_-_136918885 | 0.90 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr2_-_128890092 | 0.89 |
ENSMUST00000145798.4
|
Vinac1
|
vinculin/alpha-catenin family member 1 |
| chr6_+_41515152 | 0.88 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chr8_-_25438784 | 0.87 |
ENSMUST00000119720.8
ENSMUST00000121438.9 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr6_-_136918671 | 0.87 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr10_+_129691236 | 0.87 |
ENSMUST00000204622.3
|
Olfr813
|
olfactory receptor 813 |
| chr7_+_28050077 | 0.86 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chr3_+_69914946 | 0.84 |
ENSMUST00000053013.6
|
Otol1
|
otolin 1 |
| chr4_+_134238310 | 0.84 |
ENSMUST00000105866.3
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr15_-_74855264 | 0.84 |
ENSMUST00000023250.11
ENSMUST00000166694.2 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr3_-_98471301 | 0.83 |
ENSMUST00000058728.10
|
Gm10681
|
predicted gene 10681 |
| chr12_-_113666198 | 0.83 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr11_-_82719850 | 0.82 |
ENSMUST00000021036.13
ENSMUST00000074515.11 ENSMUST00000103218.3 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr11_-_53508160 | 0.82 |
ENSMUST00000150568.8
|
Il4
|
interleukin 4 |
| chr6_-_142453531 | 0.82 |
ENSMUST00000134191.3
ENSMUST00000239397.2 ENSMUST00000239395.2 ENSMUST00000032373.12 |
Ldhb
|
lactate dehydrogenase B |
| chr4_+_57782150 | 0.82 |
ENSMUST00000124581.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr6_+_41523664 | 0.81 |
ENSMUST00000103299.3
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr14_+_53941464 | 0.81 |
ENSMUST00000103664.6
|
Trav5-4
|
T cell receptor alpha variable 5-4 |
| chr13_+_4624074 | 0.81 |
ENSMUST00000021628.4
|
Akr1c21
|
aldo-keto reductase family 1, member C21 |
| chr7_-_102566717 | 0.81 |
ENSMUST00000214160.2
ENSMUST00000215773.2 |
Olfr571
|
olfactory receptor 571 |
| chr5_+_87973541 | 0.79 |
ENSMUST00000101056.8
ENSMUST00000002310.8 |
Prr27
|
proline rich 27 |
| chr3_+_92586546 | 0.79 |
ENSMUST00000047055.4
|
Lce1c
|
late cornified envelope 1C |
| chr12_-_81468705 | 0.79 |
ENSMUST00000085319.4
|
Adam4
|
a disintegrin and metallopeptidase domain 4 |
| chr4_+_98614971 | 0.79 |
ENSMUST00000152889.9
ENSMUST00000154279.3 |
L1td1
|
LINE-1 type transposase domain containing 1 |
| chr1_-_138102972 | 0.79 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr5_-_65248962 | 0.78 |
ENSMUST00000212080.2
|
Tmem156
|
transmembrane protein 156 |
| chr14_+_67953687 | 0.78 |
ENSMUST00000150768.8
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr5_+_110478558 | 0.77 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr2_+_173364379 | 0.77 |
ENSMUST00000029023.4
|
1700021F07Rik
|
RIKEN cDNA 1700021F07 gene |
| chr8_-_57003828 | 0.77 |
ENSMUST00000134162.8
ENSMUST00000140107.8 ENSMUST00000040330.15 ENSMUST00000135337.8 |
Cep44
|
centrosomal protein 44 |
| chr6_+_41192984 | 0.76 |
ENSMUST00000193997.6
ENSMUST00000103279.3 |
Trbv23
|
T cell receptor beta, variable V23 |
| chrY_+_52778041 | 0.76 |
ENSMUST00000178673.2
|
Gm21258
|
predicted gene, 21258 |
| chr11_-_118021460 | 0.76 |
ENSMUST00000132685.9
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr2_+_101508747 | 0.76 |
ENSMUST00000004949.8
|
Traf6
|
TNF receptor-associated factor 6 |
| chr3_+_104974160 | 0.75 |
ENSMUST00000070584.5
|
4930564D02Rik
|
RIKEN cDNA 4930564D02 gene |
| chr4_-_117035922 | 0.75 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr9_+_64080644 | 0.75 |
ENSMUST00000034966.9
|
Rpl4
|
ribosomal protein L4 |
| chrY_-_68304748 | 0.74 |
ENSMUST00000180329.2
|
Gm20937
|
predicted gene, 20937 |
| chr1_+_61017057 | 0.74 |
ENSMUST00000027162.12
ENSMUST00000102827.4 |
Icos
|
inducible T cell co-stimulator |
| chr12_-_113860566 | 0.74 |
ENSMUST00000103474.5
|
Ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr4_-_73869071 | 0.73 |
ENSMUST00000095023.2
ENSMUST00000030101.4 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr4_-_112632013 | 0.73 |
ENSMUST00000060327.4
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr16_-_19525122 | 0.73 |
ENSMUST00000081880.7
|
Lamp3
|
lysosomal-associated membrane protein 3 |
| chr6_-_54949587 | 0.73 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr10_+_84412490 | 0.73 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr17_+_87224776 | 0.73 |
ENSMUST00000042172.7
|
Tmem247
|
transmembrane protein 247 |
| chr6_-_124704237 | 0.72 |
ENSMUST00000174787.2
ENSMUST00000172690.2 |
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chrY_+_77705501 | 0.72 |
ENSMUST00000179073.2
|
Gm21650
|
predicted gene, 21650 |
| chrY_-_72256967 | 0.72 |
ENSMUST00000178505.2
|
Gm20843
|
predicted gene, 20843 |
| chr6_-_136918844 | 0.72 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr14_+_43156329 | 0.72 |
ENSMUST00000228117.2
|
Gm9732
|
predicted gene 9732 |
| chr2_-_88058678 | 0.72 |
ENSMUST00000214386.2
|
Olfr1170
|
olfactory receptor 1170 |
| chr9_-_64080161 | 0.71 |
ENSMUST00000176299.8
ENSMUST00000130127.8 ENSMUST00000176794.8 ENSMUST00000177045.8 |
Zwilch
|
zwilch kinetochore protein |
| chrY_+_80146479 | 0.71 |
ENSMUST00000179811.2
|
Gm21760
|
predicted gene, 21760 |
| chr11_+_48728291 | 0.71 |
ENSMUST00000046903.6
|
Trim7
|
tripartite motif-containing 7 |
| chr2_+_24181208 | 0.71 |
ENSMUST00000058056.2
|
Il1f10
|
interleukin 1 family, member 10 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.5 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 1.0 | 4.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.6 | 3.1 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.5 | 2.1 | GO:1904346 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.4 | 2.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.4 | 1.7 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.4 | 1.7 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.4 | 2.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.4 | 1.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.4 | 1.6 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.4 | 3.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 2.2 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.3 | 3.6 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.3 | 3.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 2.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.3 | 0.8 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.3 | 0.8 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.2 | 5.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.2 | 1.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 1.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.2 | 0.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 1.6 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 0.7 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) negative regulation of interleukin-18 production(GO:0032701) |
| 0.2 | 0.6 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.2 | 0.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 2.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 4.7 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 1.0 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.2 | 0.5 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.2 | 0.5 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.2 | 0.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 3.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 1.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 3.0 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 7.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 1.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.6 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.7 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 0.1 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.1 | 0.5 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.6 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.3 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.6 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 1.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 1.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.3 | GO:0010924 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.1 | 1.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 3.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.7 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 6.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.7 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.7 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.3 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.1 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.2 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.1 | 0.8 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.1 | 0.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.6 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.6 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 1.1 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.5 | GO:0021815 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.1 | 0.6 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.4 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 1.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.3 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 1.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.7 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 1.0 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 2.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.3 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.5 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.2 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.0 | 0.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 15.7 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.6 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.9 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 1.6 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.3 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.7 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.6 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 1.5 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.7 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.0 | 0.3 | GO:0006026 | aminoglycan catabolic process(GO:0006026) |
| 0.0 | 0.7 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.6 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 23.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.2 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 2.6 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 1.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 2.4 | GO:0048515 | spermatid differentiation(GO:0048515) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.3 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.5 | 4.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 2.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.3 | 1.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 3.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.7 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.2 | 0.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.7 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.2 | 0.7 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.2 | 1.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 1.5 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 0.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 0.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 18.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 7.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.3 | GO:0097635 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.1 | 1.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 1.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 2.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 1.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 2.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 3.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 3.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.3 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.7 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.7 | 2.0 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.6 | 3.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.6 | 2.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 2.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.5 | 1.9 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.4 | 3.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 2.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.3 | 0.7 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.3 | 1.0 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.3 | 1.7 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.3 | 2.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 3.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.3 | 0.8 | GO:0070401 | NADP+ binding(GO:0070401) lithocholic acid binding(GO:1902121) |
| 0.3 | 2.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 0.8 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 0.6 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 1.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 3.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 0.9 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 0.6 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 5.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 0.5 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 1.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.6 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 1.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 2.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.6 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 2.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 1.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.2 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.1 | 2.6 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.6 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 1.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.8 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 7.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.5 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.0 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 4.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 4.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 5.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.6 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 1.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 22.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 8.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 1.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 4.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 3.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.8 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 3.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 3.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.8 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 2.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 3.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |