Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Isl1
Z-value: 1.09
Transcription factors associated with Isl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl1
|
ENSMUSG00000042258.14 | Isl1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl1 | mm39_v1_chr13_-_116446166_116446235 | 0.13 | 2.8e-01 | Click! |
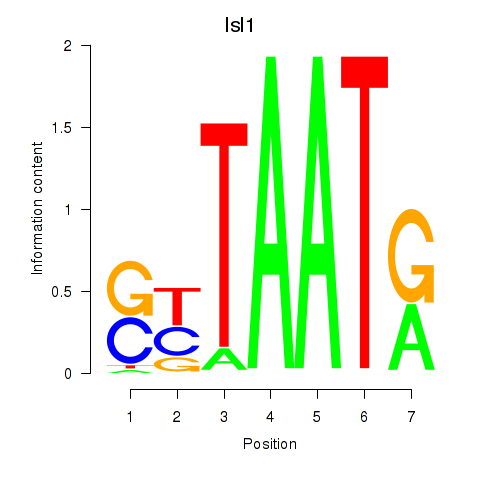
Activity profile of Isl1 motif
Sorted Z-values of Isl1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_54457978 | 6.28 |
ENSMUST00000103740.2
ENSMUST00000198398.5 |
Trac
|
T cell receptor alpha constant |
| chr12_-_113802603 | 6.02 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr17_-_52117894 | 5.71 |
ENSMUST00000176669.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr9_+_53678801 | 5.37 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr12_-_113790741 | 4.97 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr10_+_101517348 | 4.71 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chr1_-_79417732 | 4.57 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr12_-_113700190 | 4.54 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr9_-_112016966 | 4.35 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chrX_+_95498965 | 4.31 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr10_+_28544356 | 4.24 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chrX_+_162923474 | 3.96 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr1_-_173105344 | 3.83 |
ENSMUST00000111224.5
|
Mptx2
|
mucosal pentraxin 2 |
| chr17_-_78991691 | 3.81 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr11_-_76400245 | 3.62 |
ENSMUST00000094012.11
|
Abr
|
active BCR-related gene |
| chr3_+_103739877 | 3.58 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr11_-_6180127 | 3.48 |
ENSMUST00000004505.3
|
Npc1l1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr4_-_94538370 | 3.28 |
ENSMUST00000053419.9
|
Lrrc19
|
leucine rich repeat containing 19 |
| chrX_+_162873183 | 3.22 |
ENSMUST00000015545.10
|
Cltrn
|
collectrin, amino acid transport regulator |
| chr12_-_113823290 | 3.21 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr4_-_94538329 | 3.21 |
ENSMUST00000107101.2
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr1_+_172168764 | 3.19 |
ENSMUST00000056136.4
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr9_-_40257586 | 3.17 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr13_+_76727787 | 3.16 |
ENSMUST00000126960.8
ENSMUST00000109583.9 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr2_+_124994425 | 3.13 |
ENSMUST00000110494.9
ENSMUST00000110495.3 ENSMUST00000028630.9 |
Slc12a1
|
solute carrier family 12, member 1 |
| chr2_+_109522781 | 3.10 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr9_-_95697441 | 2.97 |
ENSMUST00000119760.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr19_-_8816530 | 2.96 |
ENSMUST00000096259.6
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr10_-_18890281 | 2.80 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr4_+_41903610 | 2.74 |
ENSMUST00000098128.4
|
Ccl21d
|
chemokine (C-C motif) ligand 21D |
| chr4_+_42255693 | 2.71 |
ENSMUST00000178864.3
|
Ccl21b
|
chemokine (C-C motif) ligand 21B (leucine) |
| chr18_-_47466378 | 2.70 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr4_+_42114817 | 2.67 |
ENSMUST00000098123.4
|
Gm13304
|
predicted gene 13304 |
| chr16_+_36514334 | 2.60 |
ENSMUST00000023617.13
ENSMUST00000089618.10 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr9_-_82856321 | 2.52 |
ENSMUST00000189985.2
|
Phip
|
pleckstrin homology domain interacting protein |
| chr9_+_113641615 | 2.47 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr2_+_91376650 | 2.46 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr5_-_43978980 | 2.42 |
ENSMUST00000114047.10
|
Fbxl5
|
F-box and leucine-rich repeat protein 5 |
| chr5_+_129661233 | 2.41 |
ENSMUST00000031390.10
|
Mmp17
|
matrix metallopeptidase 17 |
| chr10_+_28544556 | 2.37 |
ENSMUST00000161345.2
|
Themis
|
thymocyte selection associated |
| chr6_+_41928559 | 2.37 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr10_+_18345706 | 2.32 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr14_-_121976273 | 2.28 |
ENSMUST00000212376.3
ENSMUST00000239077.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr5_+_24679154 | 2.25 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr10_-_12490424 | 2.21 |
ENSMUST00000217994.2
|
Utrn
|
utrophin |
| chr1_-_64160557 | 2.20 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr2_-_58050494 | 2.11 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr6_+_136509922 | 2.11 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr12_-_72455708 | 2.09 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr13_-_51947685 | 2.05 |
ENSMUST00000110040.9
ENSMUST00000021900.14 |
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr10_-_108846816 | 1.99 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr9_-_112046411 | 1.95 |
ENSMUST00000161412.8
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chrX_+_56257374 | 1.95 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr9_+_37278647 | 1.94 |
ENSMUST00000051839.9
|
Hepacam
|
hepatocyte cell adhesion molecule |
| chr12_-_113666198 | 1.93 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr7_+_143792455 | 1.93 |
ENSMUST00000239495.2
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr7_+_100435548 | 1.91 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A |
| chr9_-_71803354 | 1.87 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr10_+_75983285 | 1.86 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr11_+_98277276 | 1.85 |
ENSMUST00000041301.8
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr11_+_78192355 | 1.78 |
ENSMUST00000045026.4
|
Spag5
|
sperm associated antigen 5 |
| chr14_-_78866714 | 1.73 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr5_+_20433169 | 1.67 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_+_70332836 | 1.63 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chr15_-_93417380 | 1.61 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr5_+_81169049 | 1.58 |
ENSMUST00000117253.8
ENSMUST00000120128.8 |
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr8_-_65489834 | 1.51 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr2_-_59778560 | 1.50 |
ENSMUST00000153136.2
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr3_+_109481223 | 1.50 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr3_-_7678796 | 1.50 |
ENSMUST00000192202.6
|
Il7
|
interleukin 7 |
| chr11_-_99313078 | 1.49 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr11_+_4959515 | 1.49 |
ENSMUST00000101613.3
|
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr9_+_3403592 | 1.48 |
ENSMUST00000027027.7
|
Cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr10_+_58230203 | 1.48 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr14_+_53497035 | 1.47 |
ENSMUST00000197614.2
|
Trav14n-2
|
T cell receptor alpha variable 14N-2 |
| chr3_-_144511566 | 1.43 |
ENSMUST00000199029.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr11_-_109188892 | 1.41 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr10_+_87926932 | 1.40 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr15_-_36496880 | 1.39 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr14_-_28691423 | 1.38 |
ENSMUST00000225985.2
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr14_+_53153256 | 1.36 |
ENSMUST00000197007.2
ENSMUST00000103593.3 |
Trav12d-2
|
T cell receptor alpha variable 12D-2 |
| chr14_+_10123804 | 1.34 |
ENSMUST00000022262.6
ENSMUST00000224714.2 |
Fezf2
|
Fez family zinc finger 2 |
| chr2_+_158636727 | 1.33 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr3_+_61269059 | 1.31 |
ENSMUST00000049064.4
|
Rap2b
|
RAP2B, member of RAS oncogene family |
| chr4_-_116228921 | 1.30 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr2_+_29533799 | 1.30 |
ENSMUST00000238899.2
|
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr4_+_57821050 | 1.28 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr1_+_87731360 | 1.27 |
ENSMUST00000177757.2
ENSMUST00000077772.12 |
Sag
|
S-antigen, retina and pineal gland (arrestin) |
| chr14_-_36657517 | 1.27 |
ENSMUST00000183038.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr13_-_103901010 | 1.25 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr4_+_41966058 | 1.24 |
ENSMUST00000108026.3
|
Fam205a4
|
family with sequence similarity 205, member A4 |
| chr3_-_75177378 | 1.23 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr6_+_142244145 | 1.23 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr16_-_96971905 | 1.23 |
ENSMUST00000056102.9
|
Dscam
|
DS cell adhesion molecule |
| chr5_+_107655487 | 1.23 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr5_+_44070486 | 1.19 |
ENSMUST00000122204.3
ENSMUST00000200338.2 |
Gm7879
|
predicted pseudogene 7879 |
| chr3_+_134947971 | 1.16 |
ENSMUST00000197369.3
|
Cenpe
|
centromere protein E |
| chr18_-_35348049 | 1.14 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr3_+_116388600 | 1.14 |
ENSMUST00000198386.5
ENSMUST00000198311.5 ENSMUST00000197335.2 |
Sass6
|
SAS-6 centriolar assembly protein |
| chr8_-_65489791 | 1.12 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr15_-_50746202 | 1.12 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr3_-_7678785 | 1.11 |
ENSMUST00000194279.6
|
Il7
|
interleukin 7 |
| chrX_+_113384008 | 1.11 |
ENSMUST00000113371.8
ENSMUST00000040504.12 |
Klhl4
|
kelch-like 4 |
| chr10_-_70491764 | 1.06 |
ENSMUST00000162144.2
ENSMUST00000162793.8 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr17_+_71511642 | 1.05 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr2_+_71042285 | 1.05 |
ENSMUST00000112144.9
ENSMUST00000100028.10 ENSMUST00000112136.2 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr1_-_182929025 | 1.05 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr4_+_3940747 | 1.04 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr11_+_90078485 | 1.02 |
ENSMUST00000124877.3
ENSMUST00000153734.2 ENSMUST00000134929.8 |
Gm45716
|
predicted gene 45716 |
| chr2_+_57887896 | 1.02 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr14_+_53167246 | 1.01 |
ENSMUST00000177703.3
|
Trav12d-3
|
T cell receptor alpha variable 12D-3 |
| chr18_+_37858753 | 1.00 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr1_-_58009263 | 1.00 |
ENSMUST00000114410.10
|
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr14_+_53775648 | 1.00 |
ENSMUST00000200115.2
ENSMUST00000103650.3 |
Trav12-1
|
T cell receptor alpha variable 12-1 |
| chr3_+_90383425 | 0.96 |
ENSMUST00000001042.10
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr9_-_119038162 | 0.95 |
ENSMUST00000084797.6
|
Slc22a13
|
solute carrier family 22 (organic cation transporter), member 13 |
| chr5_-_5315968 | 0.94 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr9_+_35334878 | 0.94 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr1_-_58009191 | 0.93 |
ENSMUST00000159826.2
ENSMUST00000164963.8 |
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr3_-_49711706 | 0.92 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr12_-_25147139 | 0.91 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr3_-_116388334 | 0.91 |
ENSMUST00000197190.5
ENSMUST00000198454.2 |
Trmt13
|
tRNA methyltransferase 13 |
| chr18_+_35347983 | 0.90 |
ENSMUST00000235449.2
ENSMUST00000235269.2 |
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr10_+_34359513 | 0.90 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr4_+_73931679 | 0.86 |
ENSMUST00000098006.9
ENSMUST00000084474.6 |
Frmd3
|
FERM domain containing 3 |
| chr14_+_67148619 | 0.84 |
ENSMUST00000089236.11
ENSMUST00000122431.3 |
Pnma2
|
paraneoplastic antigen MA2 |
| chr2_+_71042050 | 0.78 |
ENSMUST00000112142.8
ENSMUST00000112139.8 ENSMUST00000112140.8 ENSMUST00000112138.8 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr9_+_94551929 | 0.78 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr18_+_4375582 | 0.78 |
ENSMUST00000025077.7
|
Mtpap
|
mitochondrial poly(A) polymerase |
| chr1_+_12788720 | 0.77 |
ENSMUST00000088585.10
|
Sulf1
|
sulfatase 1 |
| chr10_+_34359395 | 0.77 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr8_-_5155347 | 0.75 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr14_-_70867588 | 0.74 |
ENSMUST00000228009.2
|
Dmtn
|
dematin actin binding protein |
| chr3_-_104725853 | 0.74 |
ENSMUST00000106775.8
ENSMUST00000166979.8 |
Mov10
|
Mov10 RISC complex RNA helicase |
| chr10_-_13264497 | 0.71 |
ENSMUST00000105546.8
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr3_+_5815863 | 0.68 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr10_+_101517556 | 0.68 |
ENSMUST00000156751.8
|
Mgat4c
|
MGAT4 family, member C |
| chr1_+_133109059 | 0.66 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr18_-_68562385 | 0.63 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr2_+_71042172 | 0.60 |
ENSMUST00000081710.12
|
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr16_-_90866032 | 0.58 |
ENSMUST00000035689.8
ENSMUST00000114076.2 |
4932438H23Rik
|
RIKEN cDNA 4932438H23 gene |
| chr5_+_20112704 | 0.57 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_+_47536075 | 0.54 |
ENSMUST00000227554.2
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr7_-_117715351 | 0.54 |
ENSMUST00000128482.8
ENSMUST00000131840.3 |
Rps15a
|
ribosomal protein S15A |
| chr13_+_76727799 | 0.52 |
ENSMUST00000109589.3
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr3_-_49711765 | 0.52 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr9_-_44679136 | 0.51 |
ENSMUST00000034607.10
|
Arcn1
|
archain 1 |
| chr17_-_71153283 | 0.50 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr6_-_129428746 | 0.48 |
ENSMUST00000204012.2
ENSMUST00000037481.10 |
Clec1a
|
C-type lectin domain family 1, member a |
| chr18_+_77032080 | 0.46 |
ENSMUST00000026485.15
ENSMUST00000150990.9 ENSMUST00000148955.3 |
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr11_-_109188917 | 0.46 |
ENSMUST00000106704.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr6_+_54794433 | 0.45 |
ENSMUST00000127331.2
|
Znrf2
|
zinc and ring finger 2 |
| chrX_-_42363663 | 0.45 |
ENSMUST00000016294.8
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr13_+_93444514 | 0.44 |
ENSMUST00000079086.8
|
Homer1
|
homer scaffolding protein 1 |
| chr4_+_150321272 | 0.43 |
ENSMUST00000080926.13
|
Eno1
|
enolase 1, alpha non-neuron |
| chr11_+_117157024 | 0.41 |
ENSMUST00000019038.15
|
Septin9
|
septin 9 |
| chr5_+_23992689 | 0.38 |
ENSMUST00000120869.6
ENSMUST00000030852.13 ENSMUST00000117783.8 ENSMUST00000115113.3 |
Rint1
|
RAD50 interactor 1 |
| chr18_+_37143758 | 0.38 |
ENSMUST00000115657.10
ENSMUST00000192447.6 |
Pcdha11
|
protocadherin alpha 11 |
| chr10_+_26648473 | 0.37 |
ENSMUST00000039557.9
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr14_+_53081922 | 0.36 |
ENSMUST00000181360.3
ENSMUST00000183652.2 |
Trav12d-1
|
T cell receptor alpha variable 12D-1 |
| chr8_-_123187406 | 0.34 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr7_-_84059170 | 0.33 |
ENSMUST00000208995.2
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr3_-_127019496 | 0.33 |
ENSMUST00000182064.9
ENSMUST00000182452.8 |
Ank2
|
ankyrin 2, brain |
| chr14_+_47535717 | 0.32 |
ENSMUST00000166743.9
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr6_-_129428869 | 0.32 |
ENSMUST00000203162.3
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr18_-_15851103 | 0.29 |
ENSMUST00000053017.13
|
Chst9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr19_+_4036562 | 0.27 |
ENSMUST00000236224.2
ENSMUST00000236510.2 ENSMUST00000237910.2 ENSMUST00000235612.2 ENSMUST00000054030.8 |
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr10_-_41894360 | 0.27 |
ENSMUST00000162405.8
ENSMUST00000095729.11 ENSMUST00000161081.2 ENSMUST00000160262.9 |
Armc2
|
armadillo repeat containing 2 |
| chrX_-_103900834 | 0.26 |
ENSMUST00000033575.7
|
Magee2
|
MAGE family member E2 |
| chr10_+_40225272 | 0.24 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr8_-_34237752 | 0.23 |
ENSMUST00000179364.3
|
Smim18
|
small integral membrane protein 18 |
| chr17_-_35454729 | 0.22 |
ENSMUST00000048994.7
|
Nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor like 1 |
| chrX_-_151820545 | 0.22 |
ENSMUST00000051484.5
|
Mageh1
|
MAGE family member H1 |
| chr19_-_40260286 | 0.22 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr18_-_42712717 | 0.21 |
ENSMUST00000054738.5
|
Gpr151
|
G protein-coupled receptor 151 |
| chr2_+_153003212 | 0.21 |
ENSMUST00000089027.3
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr4_-_137493785 | 0.20 |
ENSMUST00000139951.8
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr3_-_123484499 | 0.20 |
ENSMUST00000154668.8
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr11_-_84058292 | 0.18 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr3_-_123483772 | 0.18 |
ENSMUST00000172537.3
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr17_-_24752683 | 0.17 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr5_-_123663440 | 0.16 |
ENSMUST00000197682.6
|
Gm49027
|
predicted gene, 49027 |
| chr15_+_91722458 | 0.15 |
ENSMUST00000109277.8
|
Smgc
|
submandibular gland protein C |
| chr17_+_28945057 | 0.14 |
ENSMUST00000233003.2
|
Gm4356
|
predicted gene 4356 |
| chr2_-_120946877 | 0.14 |
ENSMUST00000110675.3
|
Tgm7
|
transglutaminase 7 |
| chr4_-_41723129 | 0.12 |
ENSMUST00000171641.2
ENSMUST00000030158.11 |
Dctn3
|
dynactin 3 |
| chrX_+_105964224 | 0.12 |
ENSMUST00000060576.8
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chrX_-_142716200 | 0.09 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr19_-_11816583 | 0.09 |
ENSMUST00000214887.2
|
Olfr1417
|
olfactory receptor 1417 |
| chr1_-_75110511 | 0.09 |
ENSMUST00000027405.6
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr2_+_67578556 | 0.08 |
ENSMUST00000180887.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr3_-_95662134 | 0.08 |
ENSMUST00000198289.5
ENSMUST00000196868.5 ENSMUST00000074339.13 ENSMUST00000163530.8 ENSMUST00000029752.15 ENSMUST00000195929.5 ENSMUST00000199570.2 ENSMUST00000098857.9 |
Tars2
|
threonyl-tRNA synthetase 2, mitochondrial (putative) |
| chr10_-_92917458 | 0.07 |
ENSMUST00000212902.2
ENSMUST00000168617.3 ENSMUST00000168110.8 ENSMUST00000020200.14 |
Cfap54
|
cilia and flagella associated protein 54 |
| chr1_+_81055201 | 0.07 |
ENSMUST00000123285.2
|
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr4_+_43406435 | 0.06 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr14_+_53497357 | 0.06 |
ENSMUST00000103623.3
|
Trav14n-2
|
T cell receptor alpha variable 14N-2 |
| chr4_+_94627513 | 0.06 |
ENSMUST00000073939.13
ENSMUST00000102798.8 |
Tek
|
TEK receptor tyrosine kinase |
| chr1_-_170695328 | 0.05 |
ENSMUST00000027974.7
|
Atf6
|
activating transcription factor 6 |
| chr2_-_17465410 | 0.04 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr16_-_21980200 | 0.02 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0061193 | taste bud development(GO:0061193) |
| 1.0 | 4.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.9 | 2.8 | GO:0002632 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.8 | 3.2 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.7 | 3.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.6 | 1.9 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.6 | 3.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.5 | 1.6 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.5 | 2.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.5 | 1.9 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.5 | 1.4 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.5 | 3.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.4 | 1.2 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.4 | 2.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 1.9 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.3 | 1.0 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.3 | 2.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.3 | 4.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.3 | 0.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.3 | 6.6 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.3 | 1.8 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.2 | 5.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 2.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 2.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 2.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 2.4 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 2.3 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 1.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 0.7 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 3.5 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 1.2 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 2.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 0.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 2.4 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.1 | 2.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 20.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 1.9 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 0.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.4 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 4.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.9 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.4 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 1.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 2.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 1.5 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 6.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.7 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 6.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 2.2 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.1 | 2.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.8 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 1.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.9 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 1.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 1.6 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 1.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 2.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 1.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 1.4 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 1.1 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 4.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 1.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 3.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 2.1 | GO:0090277 | positive regulation of peptide hormone secretion(GO:0090277) |
| 0.0 | 0.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 4.8 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 1.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.3 | 3.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 6.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 2.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 0.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 20.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 3.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 1.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 3.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 3.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 6.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 4.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 1.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 4.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 3.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.8 | 3.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.7 | 2.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.7 | 4.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.7 | 2.6 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.6 | 1.9 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.5 | 5.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 3.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 4.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 2.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 1.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 2.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 4.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 4.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 3.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 1.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 20.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 3.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 4.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.6 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 3.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 2.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.0 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.3 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.2 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 0.4 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 6.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 6.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 5.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 1.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.9 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 1.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 6.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.3 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 6.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.7 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 4.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 5.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 3.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 3.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 3.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 5.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 6.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 4.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 5.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 5.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 5.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 2.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |