Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Isl2
Z-value: 0.83
Transcription factors associated with Isl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl2
|
ENSMUSG00000032318.13 | Isl2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl2 | mm39_v1_chr9_+_55448432_55448493 | 0.13 | 2.9e-01 | Click! |
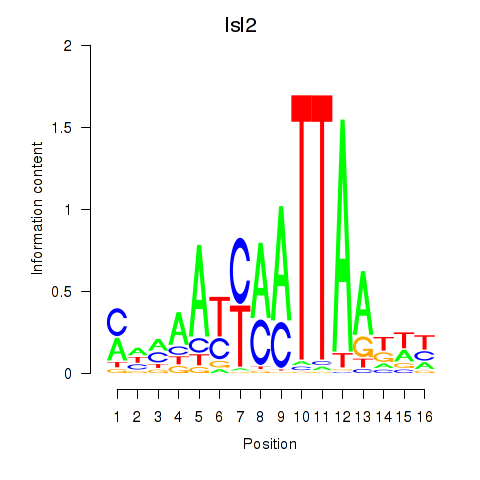
Activity profile of Isl2 motif
Sorted Z-values of Isl2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_78347636 | 12.74 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr6_-_69282389 | 8.04 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr3_-_72875187 | 7.44 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr6_+_68279392 | 7.24 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr6_-_69584812 | 6.86 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr6_+_67838100 | 6.16 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr8_+_21999274 | 6.12 |
ENSMUST00000084042.4
|
Defa20
|
defensin, alpha, 20 |
| chr8_+_21691577 | 5.94 |
ENSMUST00000110754.2
|
Defa41
|
defensin, alpha, 41 |
| chr1_-_82746169 | 5.38 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr6_-_69204417 | 5.12 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr15_-_58261093 | 4.70 |
ENSMUST00000227274.3
|
Anxa13
|
annexin A13 |
| chr10_-_75946790 | 4.47 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr17_-_28779678 | 4.47 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr12_-_114487525 | 4.46 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr5_-_44139099 | 4.38 |
ENSMUST00000061299.9
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr5_-_44139121 | 4.23 |
ENSMUST00000199894.2
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr12_-_113802603 | 3.87 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr8_-_65582206 | 3.82 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr6_-_41012435 | 3.80 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr10_+_101517348 | 3.65 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chr6_-_129077867 | 3.59 |
ENSMUST00000032258.8
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr6_-_68746087 | 3.56 |
ENSMUST00000103333.4
|
Igkv4-91
|
immunoglobulin kappa chain variable 4-91 |
| chr10_+_88721854 | 3.48 |
ENSMUST00000020255.8
|
Slc5a8
|
solute carrier family 5 (iodide transporter), member 8 |
| chr10_+_115979787 | 3.45 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr9_+_53678801 | 3.42 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr6_-_70313491 | 3.39 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chrX_-_133012457 | 3.32 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chrX_+_95498965 | 3.03 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr16_+_45044678 | 2.88 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr19_-_24178000 | 2.81 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr12_-_115832846 | 2.71 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr6_-_70116066 | 2.69 |
ENSMUST00000103379.3
ENSMUST00000197371.2 |
Igkv6-29
|
immunoglobulin kappa chain variable 6-29 |
| chr2_+_57887896 | 2.66 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr10_+_23672842 | 2.60 |
ENSMUST00000119597.8
ENSMUST00000179321.8 ENSMUST00000133289.2 |
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr12_-_113649535 | 2.60 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr14_+_51366512 | 2.58 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr11_-_80670815 | 2.54 |
ENSMUST00000041065.14
ENSMUST00000070997.6 |
Myo1d
|
myosin ID |
| chr12_-_114443071 | 2.53 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr6_+_40619913 | 2.49 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr4_-_111755892 | 2.35 |
ENSMUST00000102720.8
|
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr5_-_140986312 | 2.31 |
ENSMUST00000085786.7
|
Card11
|
caspase recruitment domain family, member 11 |
| chr17_+_12338161 | 2.30 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr2_+_24257576 | 2.25 |
ENSMUST00000140547.2
ENSMUST00000102942.8 |
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chrX_+_72719098 | 2.24 |
ENSMUST00000171398.2
|
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr10_+_94412116 | 2.16 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr4_+_135899678 | 2.12 |
ENSMUST00000061721.6
|
E2f2
|
E2F transcription factor 2 |
| chr3_-_67371161 | 2.05 |
ENSMUST00000058981.3
|
Lxn
|
latexin |
| chr3_-_9069514 | 2.04 |
ENSMUST00000091355.12
ENSMUST00000134788.8 |
Tpd52
|
tumor protein D52 |
| chr6_+_78382131 | 2.03 |
ENSMUST00000023906.4
|
Reg2
|
regenerating islet-derived 2 |
| chr6_-_69020489 | 2.02 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chrX_-_133012600 | 1.76 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr5_-_105198913 | 1.75 |
ENSMUST00000112718.5
|
Gbp8
|
guanylate-binding protein 8 |
| chrX_-_73472477 | 1.69 |
ENSMUST00000143521.2
|
G6pdx
|
glucose-6-phosphate dehydrogenase X-linked |
| chr19_-_38031774 | 1.68 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr6_-_113411718 | 1.66 |
ENSMUST00000113091.8
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr6_-_69162381 | 1.64 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr6_+_29859685 | 1.63 |
ENSMUST00000134438.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr10_-_84963841 | 1.62 |
ENSMUST00000214193.2
ENSMUST00000050813.4 ENSMUST00000217027.2 |
Mterf2
|
mitochondrial transcription termination factor 2 |
| chr10_+_101994719 | 1.62 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr13_-_19521337 | 1.60 |
ENSMUST00000103563.3
|
Trgv2
|
T cell receptor gamma variable 2 |
| chr2_+_68966125 | 1.58 |
ENSMUST00000041865.8
|
Nostrin
|
nitric oxide synthase trafficker |
| chr6_-_3399451 | 1.54 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr1_-_153061758 | 1.52 |
ENSMUST00000185356.7
|
Lamc2
|
laminin, gamma 2 |
| chr1_+_156666485 | 1.48 |
ENSMUST00000111720.2
|
Angptl1
|
angiopoietin-like 1 |
| chr7_+_126575510 | 1.46 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr1_-_57008986 | 1.41 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chrX_+_163221035 | 1.40 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr4_+_3940747 | 1.40 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr10_+_79832313 | 1.34 |
ENSMUST00000132517.8
|
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr6_+_67873135 | 1.32 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr13_-_21330037 | 1.29 |
ENSMUST00000216039.3
|
Olfr1368
|
olfactory receptor 1368 |
| chr16_-_19241884 | 1.24 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chr6_+_65567373 | 1.24 |
ENSMUST00000114236.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr18_+_23886765 | 1.23 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr14_-_104760051 | 1.23 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr2_+_109111083 | 1.21 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr3_-_75177378 | 1.21 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr1_-_79838897 | 1.21 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr11_+_46345784 | 1.20 |
ENSMUST00000109229.2
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr6_+_41515152 | 1.19 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chr10_+_101517556 | 1.18 |
ENSMUST00000156751.8
|
Mgat4c
|
MGAT4 family, member C |
| chr12_-_114012399 | 1.16 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr19_+_45064539 | 1.14 |
ENSMUST00000026234.5
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chr19_+_5540483 | 1.11 |
ENSMUST00000209469.2
ENSMUST00000116560.3 |
Cfl1
|
cofilin 1, non-muscle |
| chr3_-_86827664 | 1.11 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr5_+_73164226 | 1.10 |
ENSMUST00000031127.11
ENSMUST00000201304.2 |
Slc10a4
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 4 |
| chr9_-_22171180 | 1.09 |
ENSMUST00000086281.5
|
Zfp599
|
zinc finger protein 599 |
| chrY_-_1245685 | 1.06 |
ENSMUST00000143286.8
ENSMUST00000137048.8 ENSMUST00000069309.14 ENSMUST00000139365.8 ENSMUST00000154004.8 |
Uty
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr6_-_69377081 | 1.06 |
ENSMUST00000177795.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr17_+_14087827 | 1.05 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr7_+_89814713 | 1.04 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr10_-_13744676 | 1.03 |
ENSMUST00000019942.6
ENSMUST00000162610.8 |
Aig1
|
androgen-induced 1 |
| chr16_-_44153288 | 1.02 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr16_-_48592319 | 1.01 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chrX_+_162694397 | 1.01 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chrX_+_135145813 | 1.01 |
ENSMUST00000048687.11
|
Tceal9
|
transcription elongation factor A like 9 |
| chr6_-_130208601 | 1.01 |
ENSMUST00000088011.11
ENSMUST00000112013.8 ENSMUST00000049304.14 |
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr6_-_136899167 | 1.00 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr16_-_44153498 | 0.98 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr10_-_80754016 | 0.95 |
ENSMUST00000057623.14
ENSMUST00000179022.8 |
Lmnb2
|
lamin B2 |
| chr14_+_76714350 | 0.95 |
ENSMUST00000140251.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr1_-_169938060 | 0.94 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr9_+_107765320 | 0.94 |
ENSMUST00000191906.6
ENSMUST00000035202.4 |
Mon1a
|
MON1 homolog A, secretory traffciking associated |
| chr6_-_69220672 | 0.91 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr6_+_129554868 | 0.90 |
ENSMUST00000053708.9
|
Klre1
|
killer cell lectin-like receptor family E member 1 |
| chr6_-_136918885 | 0.87 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr1_-_144427302 | 0.86 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr19_+_5540591 | 0.85 |
ENSMUST00000237122.2
|
Cfl1
|
cofilin 1, non-muscle |
| chr4_+_145311759 | 0.85 |
ENSMUST00000119718.8
|
Zfp268
|
zinc finger protein 268 |
| chr7_+_28580847 | 0.85 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr7_+_126575752 | 0.84 |
ENSMUST00000206346.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr1_+_12788720 | 0.84 |
ENSMUST00000088585.10
|
Sulf1
|
sulfatase 1 |
| chr2_-_89408791 | 0.84 |
ENSMUST00000217402.2
|
Olfr1245
|
olfactory receptor 1245 |
| chr9_+_96140781 | 0.83 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr18_-_73887528 | 0.82 |
ENSMUST00000041138.3
|
Elac1
|
elaC ribonuclease Z 1 |
| chr2_-_20948230 | 0.81 |
ENSMUST00000140230.2
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr11_-_115918784 | 0.81 |
ENSMUST00000106454.8
|
H3f3b
|
H3.3 histone B |
| chr11_-_101169753 | 0.80 |
ENSMUST00000168089.2
ENSMUST00000017332.4 |
Coa3
|
cytochrome C oxidase assembly factor 3 |
| chr18_+_77877611 | 0.79 |
ENSMUST00000238172.2
|
Pstpip2
|
proline-serine-threonine phosphatase-interacting protein 2 |
| chr1_-_169938298 | 0.79 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr14_+_51366306 | 0.79 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr4_+_32623985 | 0.78 |
ENSMUST00000108178.2
|
Casp8ap2
|
caspase 8 associated protein 2 |
| chr13_-_78344492 | 0.76 |
ENSMUST00000125176.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr13_-_22289994 | 0.76 |
ENSMUST00000227357.2
ENSMUST00000228428.2 |
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr8_+_88845375 | 0.75 |
ENSMUST00000095214.10
|
Cnep1r1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr11_-_82799186 | 0.74 |
ENSMUST00000103213.10
|
Nle1
|
notchless homolog 1 |
| chr12_-_84664001 | 0.74 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr9_+_96140750 | 0.72 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr9_-_51240201 | 0.72 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr19_+_29929208 | 0.71 |
ENSMUST00000136850.2
|
Il33
|
interleukin 33 |
| chr12_+_102094977 | 0.70 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr1_-_165462020 | 0.70 |
ENSMUST00000194437.6
ENSMUST00000068705.13 ENSMUST00000111435.9 ENSMUST00000193023.2 |
Mpzl1
|
myelin protein zero-like 1 |
| chr16_-_48592372 | 0.69 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr2_+_85648823 | 0.68 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr7_+_126575781 | 0.67 |
ENSMUST00000206450.2
ENSMUST00000205830.2 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr9_-_100388857 | 0.67 |
ENSMUST00000112874.4
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chr4_-_43710231 | 0.66 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr19_-_12313274 | 0.63 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr9_-_119170456 | 0.62 |
ENSMUST00000139870.2
|
Myd88
|
myeloid differentiation primary response gene 88 |
| chr8_-_112064783 | 0.61 |
ENSMUST00000056157.14
ENSMUST00000120432.3 |
Mlkl
|
mixed lineage kinase domain-like |
| chr6_-_115014777 | 0.61 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr2_+_163500290 | 0.61 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr7_+_108265625 | 0.58 |
ENSMUST00000213979.3
ENSMUST00000216331.2 ENSMUST00000217170.2 |
Olfr510
|
olfactory receptor 510 |
| chr18_+_44237474 | 0.58 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr6_-_115569504 | 0.57 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr10_-_78554104 | 0.57 |
ENSMUST00000005488.9
|
Casp14
|
caspase 14 |
| chr13_+_104365880 | 0.57 |
ENSMUST00000022227.8
|
Cenpk
|
centromere protein K |
| chr17_+_37361523 | 0.57 |
ENSMUST00000172792.8
ENSMUST00000174347.2 |
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr10_+_129153986 | 0.56 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr15_-_37459570 | 0.56 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chr14_-_68771138 | 0.55 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chrX_+_100492684 | 0.55 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr11_-_73382303 | 0.54 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr14_+_59716265 | 0.54 |
ENSMUST00000224893.2
|
Cab39l
|
calcium binding protein 39-like |
| chrM_+_8603 | 0.53 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr4_-_88640691 | 0.53 |
ENSMUST00000094993.3
|
Klhl9
|
kelch-like 9 |
| chr9_-_113537277 | 0.53 |
ENSMUST00000111861.4
ENSMUST00000035086.13 |
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chr3_-_49711706 | 0.53 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr12_+_119407145 | 0.53 |
ENSMUST00000048880.7
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr8_-_106223502 | 0.52 |
ENSMUST00000212303.2
|
Zdhhc1
|
zinc finger, DHHC domain containing 1 |
| chr2_-_109111064 | 0.51 |
ENSMUST00000147770.2
|
Mettl15
|
methyltransferase like 15 |
| chr19_+_5524701 | 0.51 |
ENSMUST00000165485.8
ENSMUST00000166253.8 ENSMUST00000167371.8 ENSMUST00000167855.8 ENSMUST00000070118.14 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr6_-_129599645 | 0.50 |
ENSMUST00000032252.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr9_+_35334878 | 0.50 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr2_+_127696548 | 0.50 |
ENSMUST00000028859.8
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr3_-_86827640 | 0.49 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr3_+_7494108 | 0.49 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr1_+_82817794 | 0.48 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr8_+_88845406 | 0.48 |
ENSMUST00000121097.8
ENSMUST00000117775.2 |
Cnep1r1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr17_-_56312555 | 0.47 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr10_+_129493563 | 0.47 |
ENSMUST00000217094.2
|
Olfr800
|
olfactory receptor 800 |
| chr10_+_29074950 | 0.47 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr3_+_87343127 | 0.46 |
ENSMUST00000166297.7
ENSMUST00000049926.15 ENSMUST00000178261.3 |
Fcrl5
|
Fc receptor-like 5 |
| chr7_-_104246386 | 0.46 |
ENSMUST00000057385.5
|
Olfr655
|
olfactory receptor 655 |
| chr7_+_19927635 | 0.46 |
ENSMUST00000168984.2
|
Vmn1r95
|
vomeronasal 1 receptor, 95 |
| chr3_-_49711765 | 0.46 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr1_+_5658716 | 0.45 |
ENSMUST00000160777.8
ENSMUST00000239100.2 ENSMUST00000027038.11 |
Oprk1
|
opioid receptor, kappa 1 |
| chrX_+_9751861 | 0.44 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr6_-_69609162 | 0.44 |
ENSMUST00000199437.2
|
Igkv4-54
|
immunoglobulin kappa chain variable 4-54 |
| chr16_-_17379103 | 0.44 |
ENSMUST00000232637.2
ENSMUST00000100123.10 ENSMUST00000023442.13 |
Lrrc74b
|
leucine rich repeat containing 74B |
| chr2_-_88531671 | 0.43 |
ENSMUST00000104892.2
|
Olfr1196
|
olfactory receptor 1196 |
| chr10_-_129710624 | 0.43 |
ENSMUST00000081367.2
|
Olfr814
|
olfactory receptor 814 |
| chr15_+_25774070 | 0.43 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr15_+_65682066 | 0.42 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr9_+_21634779 | 0.41 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr7_+_86175967 | 0.41 |
ENSMUST00000071112.4
|
Olfr297
|
olfactory receptor 297 |
| chr11_+_67167950 | 0.41 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr8_+_96429665 | 0.40 |
ENSMUST00000073139.14
ENSMUST00000080666.8 ENSMUST00000212160.2 |
Ndrg4
|
N-myc downstream regulated gene 4 |
| chr10_-_129000620 | 0.40 |
ENSMUST00000214271.2
|
Olfr771
|
olfactory receptor 771 |
| chr9_+_58536386 | 0.39 |
ENSMUST00000176250.2
|
Nptn
|
neuroplastin |
| chr12_-_118930130 | 0.39 |
ENSMUST00000035515.5
|
Abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr2_+_89642395 | 0.38 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr18_+_44237577 | 0.37 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr2_-_150097511 | 0.37 |
ENSMUST00000063463.6
|
Gm21994
|
predicted gene 21994 |
| chr17_+_17622934 | 0.36 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr2_+_87609827 | 0.36 |
ENSMUST00000105210.3
|
Olfr152
|
olfactory receptor 152 |
| chr17_+_18518361 | 0.36 |
ENSMUST00000231938.2
ENSMUST00000079206.8 ENSMUST00000231879.3 |
Vmn2r93
|
vomeronasal 2, receptor 93 |
| chr6_-_129449739 | 0.33 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr16_-_20972750 | 0.33 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr9_+_56326089 | 0.33 |
ENSMUST00000213242.2
ENSMUST00000214771.2 ENSMUST00000217518.2 ENSMUST00000214869.2 |
Hmg20a
|
high mobility group 20A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 1.0 | 12.7 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.9 | 8.6 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.7 | 2.2 | GO:0015881 | creatine transport(GO:0015881) |
| 0.7 | 2.0 | GO:2000769 | positive regulation of NMDA glutamate receptor activity(GO:1904783) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.6 | 3.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.6 | 3.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.6 | 1.7 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.5 | 1.6 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.5 | 2.1 | GO:0002856 | negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.5 | 6.8 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.4 | 1.3 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.4 | 1.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.4 | 2.8 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.3 | 1.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 2.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.3 | 9.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 1.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 2.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 1.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 49.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 2.9 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 1.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 1.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 0.5 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.2 | 0.7 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 0.8 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 0.8 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 0.6 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.2 | 1.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.4 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.1 | 0.8 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.8 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.8 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 18.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.4 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.1 | 1.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.7 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 1.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.4 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.9 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 1.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.5 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.1 | 1.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 4.5 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 3.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.6 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.1 | 0.6 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.5 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 0.6 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 1.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.6 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.8 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 1.6 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.8 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 2.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 1.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 1.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 1.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 1.7 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 2.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.4 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:2000911 | positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 1.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.5 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.9 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.6 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 6.2 | GO:0045861 | negative regulation of proteolysis(GO:0045861) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 3.5 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 2.2 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.5 | 1.5 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.4 | 1.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.3 | 12.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 1.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 18.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 4.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 1.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 2.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 63.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 5.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 2.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 16.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 3.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 5.1 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 1.0 | 3.0 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.8 | 5.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.8 | 2.5 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.7 | 2.2 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.6 | 6.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.6 | 6.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.4 | 1.3 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.4 | 4.7 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.3 | 1.7 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.3 | 2.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 3.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.6 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 8.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 0.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 2.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 0.5 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.2 | 1.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.6 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.2 | 1.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 18.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 2.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 2.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.5 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 2.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.7 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 13.0 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 1.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 6.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 2.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.4 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 1.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 4.4 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 3.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.9 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 2.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 10.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 4.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 6.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 4.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 4.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 3.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 0.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 2.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 3.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.9 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 2.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 3.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 2.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |