Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
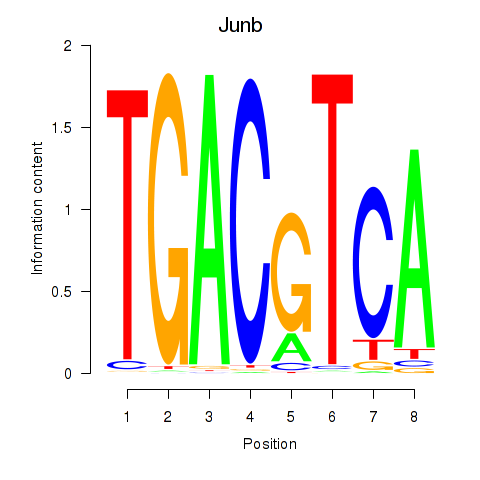
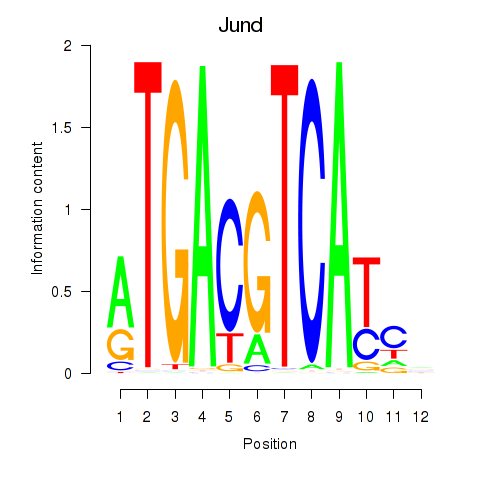
Results for Junb_Jund
Z-value: 3.05
Transcription factors associated with Junb_Jund
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Junb
|
ENSMUSG00000052837.8 | Junb |
|
Jund
|
ENSMUSG00000071076.9 | Jund |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Jund | mm39_v1_chr8_+_71151581_71151599 | 0.11 | 3.7e-01 | Click! |
| Junb | mm39_v1_chr8_-_85705338_85705355 | -0.08 | 4.9e-01 | Click! |
Activity profile of Junb_Jund motif
Sorted Z-values of Junb_Jund motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Junb_Jund
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_155118217 | 52.10 |
ENSMUST00000029128.4
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr17_-_46798566 | 39.96 |
ENSMUST00000047034.9
|
Ttbk1
|
tau tubulin kinase 1 |
| chr12_+_4132567 | 39.60 |
ENSMUST00000020986.15
ENSMUST00000049584.6 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr6_-_139987135 | 37.74 |
ENSMUST00000032356.13
|
Plcz1
|
phospholipase C, zeta 1 |
| chr19_-_5135510 | 36.54 |
ENSMUST00000140389.8
ENSMUST00000151413.2 ENSMUST00000077066.8 |
Tmem151a
|
transmembrane protein 151A |
| chr6_+_139987275 | 36.27 |
ENSMUST00000043797.6
|
Capza3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr6_+_126830102 | 31.00 |
ENSMUST00000202878.4
ENSMUST00000202574.2 |
Akap3
|
A kinase (PRKA) anchor protein 3 |
| chr9_-_106438798 | 30.46 |
ENSMUST00010126732.2
ENSMUST00010126033.2 ENSMUST00010181659.1 ENSMUST00010126065.2 ENSMUST00010126032.3 ENSMUST00000062917.16 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr17_+_33651864 | 29.28 |
ENSMUST00000174088.3
|
Actl9
|
actin-like 9 |
| chr4_-_120966396 | 28.68 |
ENSMUST00000106268.4
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chr12_-_64521464 | 28.45 |
ENSMUST00000059833.8
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr4_-_128154709 | 27.93 |
ENSMUST00000053830.5
|
Hmgb4
|
high-mobility group box 4 |
| chr6_+_126830050 | 27.10 |
ENSMUST00000095440.9
|
Akap3
|
A kinase (PRKA) anchor protein 3 |
| chr14_+_52155874 | 26.86 |
ENSMUST00000008957.13
|
Tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr16_-_16647139 | 26.60 |
ENSMUST00000023468.6
|
Spag6l
|
sperm associated antigen 6-like |
| chr7_-_103792462 | 26.56 |
ENSMUST00000057254.6
|
Ubqln3
|
ubiquilin 3 |
| chr9_-_106448182 | 26.37 |
ENSMUST00000085111.5
|
Iqcf4
|
IQ motif containing F4 |
| chr12_-_28632514 | 25.36 |
ENSMUST00000110917.2
ENSMUST00000020965.14 |
Allc
|
allantoicase |
| chr11_-_23469181 | 25.26 |
ENSMUST00000239488.2
ENSMUST00000020527.13 |
1700093K21Rik
|
RIKEN cDNA 1700093K21 gene |
| chr4_-_58009118 | 24.67 |
ENSMUST00000102897.11
ENSMUST00000239406.2 |
Txndc8
|
thioredoxin domain containing 8 |
| chr1_-_56676589 | 24.41 |
ENSMUST00000062085.6
|
Hsfy2
|
heat shock transcription factor, Y-linked 2 |
| chr4_+_56743407 | 24.16 |
ENSMUST00000095079.6
|
Actl7a
|
actin-like 7a |
| chr1_+_170136372 | 24.13 |
ENSMUST00000056991.6
|
Spata46
|
spermatogenesis associated 46 |
| chr6_+_29319132 | 24.07 |
ENSMUST00000090487.12
ENSMUST00000164560.8 |
Fam71f1
|
family with sequence similarity 71, member F1 |
| chr6_-_135231324 | 23.86 |
ENSMUST00000111911.9
ENSMUST00000111910.4 |
Gsg1
|
germ cell associated 1 |
| chr2_+_143874979 | 23.72 |
ENSMUST00000037722.9
ENSMUST00000110032.2 |
Banf2
|
BANF family member 2 |
| chr4_+_43983472 | 23.62 |
ENSMUST00000095107.3
|
Ccin
|
calicin |
| chr9_+_106391771 | 23.57 |
ENSMUST00000085113.5
|
Iqcf5
|
IQ motif containing F5 |
| chr3_+_67281424 | 23.27 |
ENSMUST00000077916.12
|
Mlf1
|
myeloid leukemia factor 1 |
| chr3_-_92050043 | 23.00 |
ENSMUST00000197811.2
ENSMUST00000029535.6 |
4930511M18Rik
Lelp1
|
RIKEN cDNA 4930511M18 gene late cornified envelope-like proline-rich 1 |
| chr7_-_24134919 | 22.83 |
ENSMUST00000080594.8
|
Irgc1
|
immunity-related GTPase family, cinema 1 |
| chr6_+_29281134 | 22.70 |
ENSMUST00000115286.4
|
Fam71f2
|
family with sequence similarity 71, member F2 |
| chr4_-_63540653 | 22.68 |
ENSMUST00000102861.8
ENSMUST00000102862.4 |
Tex48
|
testis expressed 48 |
| chr7_+_45271229 | 22.21 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr11_+_62842019 | 22.03 |
ENSMUST00000035854.4
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr9_+_106377181 | 21.91 |
ENSMUST00000085114.8
|
Iqcf1
|
IQ motif containing F1 |
| chr1_+_86354045 | 21.81 |
ENSMUST00000046004.6
|
Tex44
|
testis expressed 44 |
| chr3_-_107462378 | 21.66 |
ENSMUST00000052853.8
|
Ubl4b
|
ubiquitin-like 4B |
| chr16_+_17712061 | 21.51 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr3_+_67281449 | 21.47 |
ENSMUST00000061322.10
|
Mlf1
|
myeloid leukemia factor 1 |
| chr3_-_124374723 | 21.40 |
ENSMUST00000180162.8
ENSMUST00000047110.14 ENSMUST00000178485.8 |
1700003H04Rik
|
RIKEN cDNA 1700003H04 gene |
| chr8_-_13612397 | 21.30 |
ENSMUST00000187391.7
ENSMUST00000134023.9 ENSMUST00000151400.10 |
1700029H14Rik
|
RIKEN cDNA 1700029H14 gene |
| chr8_+_12623016 | 21.30 |
ENSMUST00000210276.2
ENSMUST00000010579.8 ENSMUST00000209428.2 |
Spaca7
|
sperm acrosome associated 7 |
| chr4_+_51216645 | 21.21 |
ENSMUST00000166749.2
ENSMUST00000156384.4 |
Cylc2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr16_-_10608766 | 21.09 |
ENSMUST00000050864.7
|
Prm3
|
protamine 3 |
| chr9_-_106421834 | 21.03 |
ENSMUST00010181660.1
ENSMUST00000215525.2 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr4_-_41517326 | 21.02 |
ENSMUST00000030152.13
ENSMUST00000095126.5 |
1110017D15Rik
|
RIKEN cDNA 1110017D15 gene |
| chr18_+_50411431 | 20.77 |
ENSMUST00000039121.4
ENSMUST00000238078.2 |
Fam170a
|
family with sequence similarity 170, member A |
| chr8_+_94537460 | 20.51 |
ENSMUST00000034198.15
ENSMUST00000125716.8 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr5_+_25386452 | 20.46 |
ENSMUST00000030778.9
|
Galntl5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr16_+_20514925 | 20.42 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr14_+_96118660 | 20.42 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr1_+_75456173 | 20.06 |
ENSMUST00000113575.9
ENSMUST00000148980.2 ENSMUST00000050899.7 ENSMUST00000187411.2 |
Tmem198
|
transmembrane protein 198 |
| chr9_+_106377153 | 19.94 |
ENSMUST00000164965.3
|
Iqcf1
|
IQ motif containing F1 |
| chr11_+_101877876 | 19.93 |
ENSMUST00000010985.8
|
Cfap97d1
|
CFAP97 domain containing 1 |
| chr19_+_8718073 | 19.91 |
ENSMUST00000163172.2
|
1700092M07Rik
|
RIKEN cDNA 1700092M07 gene |
| chr2_+_172314433 | 19.72 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209 |
| chr7_-_30443106 | 19.72 |
ENSMUST00000182634.8
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr2_+_18703797 | 19.67 |
ENSMUST00000095132.10
|
Spag6
|
sperm associated antigen 6 |
| chr11_+_108573428 | 19.57 |
ENSMUST00000106718.10
ENSMUST00000106715.8 ENSMUST00000106724.10 |
Cep112
|
centrosomal protein 112 |
| chrX_+_110154017 | 19.38 |
ENSMUST00000210720.3
|
Cylc1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr6_+_29319190 | 19.30 |
ENSMUST00000166462.2
|
Fam71f1
|
family with sequence similarity 71, member F1 |
| chr12_-_110649040 | 19.17 |
ENSMUST00000222915.2
ENSMUST00000070659.7 |
1700001K19Rik
|
RIKEN cDNA 1700001K19 gene |
| chr6_-_124441731 | 19.06 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr1_-_160986880 | 18.80 |
ENSMUST00000135643.8
ENSMUST00000178511.3 |
Tex50
|
testis expressed 50 |
| chr14_-_75830550 | 18.75 |
ENSMUST00000164082.9
ENSMUST00000169658.9 |
Cby2
|
chibby family member 2 |
| chr5_+_25386487 | 18.52 |
ENSMUST00000114965.2
|
Galntl5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr1_-_75455915 | 18.05 |
ENSMUST00000079205.14
ENSMUST00000094818.4 |
Chpf
|
chondroitin polymerizing factor |
| chrX_+_9715942 | 17.79 |
ENSMUST00000057113.3
|
H2al3
|
H2A histone family member L3 |
| chr6_-_135231168 | 17.76 |
ENSMUST00000111909.8
|
Gsg1
|
germ cell associated 1 |
| chr7_+_44145987 | 17.67 |
ENSMUST00000107927.5
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr14_-_101437750 | 17.50 |
ENSMUST00000187304.2
|
Prr30
|
proline rich 30 |
| chr15_-_99185050 | 17.46 |
ENSMUST00000109100.2
|
Fam186b
|
family with sequence similarity 186, member B |
| chr3_+_19562771 | 17.45 |
ENSMUST00000029132.11
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr11_+_70506674 | 17.27 |
ENSMUST00000180052.8
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr4_-_94817056 | 17.09 |
ENSMUST00000107097.9
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr3_+_19562753 | 17.06 |
ENSMUST00000118968.8
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr11_+_115331365 | 17.05 |
ENSMUST00000093914.5
|
Trim80
|
tripartite motif-containing 80 |
| chr14_+_66581818 | 16.99 |
ENSMUST00000118426.8
ENSMUST00000121955.8 ENSMUST00000120229.8 ENSMUST00000134440.2 |
Stmn4
|
stathmin-like 4 |
| chr3_+_153549846 | 16.99 |
ENSMUST00000044089.4
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr7_-_103778992 | 16.95 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chrX_+_165127688 | 16.95 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr2_+_18703863 | 16.64 |
ENSMUST00000173763.2
|
Spag6
|
sperm associated antigen 6 |
| chr2_+_49509288 | 16.63 |
ENSMUST00000028102.14
|
Kif5c
|
kinesin family member 5C |
| chr1_-_33946802 | 16.62 |
ENSMUST00000115161.8
ENSMUST00000129464.8 ENSMUST00000062289.11 |
Bend6
|
BEN domain containing 6 |
| chr11_+_70506716 | 16.58 |
ENSMUST00000144960.2
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr17_+_48400153 | 16.52 |
ENSMUST00000233043.2
|
1700067P10Rik
|
RIKEN cDNA 1700067P10 gene |
| chr13_-_99653045 | 16.46 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr4_-_94817025 | 16.46 |
ENSMUST00000030309.6
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr7_-_4774277 | 16.32 |
ENSMUST00000174409.2
|
Fam71e2
|
family with sequence similarity 71, member E2 |
| chr15_+_81820954 | 16.29 |
ENSMUST00000038757.8
ENSMUST00000230633.2 |
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr11_-_118233326 | 16.09 |
ENSMUST00000103024.4
|
Cep295nl
|
CEP295 N-terminal like |
| chr14_+_70815250 | 16.00 |
ENSMUST00000228554.2
|
Nudt18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr17_-_57181420 | 15.98 |
ENSMUST00000043062.5
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr4_-_25242858 | 15.87 |
ENSMUST00000029922.14
ENSMUST00000108204.2 |
Fhl5
|
four and a half LIM domains 5 |
| chr11_+_70350725 | 15.70 |
ENSMUST00000147289.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr4_-_56741398 | 15.66 |
ENSMUST00000095080.5
|
Actl7b
|
actin-like 7b |
| chr1_-_156936197 | 15.52 |
ENSMUST00000187546.7
ENSMUST00000118207.8 ENSMUST00000027884.13 ENSMUST00000121911.8 |
Tex35
|
testis expressed 35 |
| chr3_-_145813802 | 15.39 |
ENSMUST00000160285.2
|
Dnai3
|
dynein axonemal intermediate chain 3 |
| chr11_-_103529678 | 15.37 |
ENSMUST00000107014.8
ENSMUST00000021328.8 |
Lyzl6
|
lysozyme-like 6 |
| chr11_+_70350963 | 15.36 |
ENSMUST00000126105.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr5_+_101912939 | 15.34 |
ENSMUST00000031273.9
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr10_+_100428246 | 15.12 |
ENSMUST00000041162.13
ENSMUST00000190386.7 ENSMUST00000190708.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr11_+_70350436 | 15.10 |
ENSMUST00000039093.10
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr19_+_5385672 | 14.96 |
ENSMUST00000043380.5
|
Catsper1
|
cation channel, sperm associated 1 |
| chr2_-_79738773 | 14.95 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_-_126430636 | 14.93 |
ENSMUST00000205320.2
ENSMUST00000061695.5 |
4930451I11Rik
|
RIKEN cDNA 4930451I11 gene |
| chr7_-_38227617 | 14.83 |
ENSMUST00000079759.6
|
Gm5591
|
predicted gene 5591 |
| chr5_-_82272549 | 14.74 |
ENSMUST00000188072.2
ENSMUST00000185410.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr10_-_82152373 | 14.66 |
ENSMUST00000217661.2
|
4932415D10Rik
|
RIKEN cDNA 4932415D10 gene |
| chr18_-_23174698 | 14.63 |
ENSMUST00000097651.10
|
Nol4
|
nucleolar protein 4 |
| chr15_-_33687986 | 14.59 |
ENSMUST00000042021.5
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr11_+_87295860 | 14.45 |
ENSMUST00000060835.12
|
Tex14
|
testis expressed gene 14 |
| chrX_+_117336912 | 14.23 |
ENSMUST00000072518.8
|
Tgif2lx2
|
TGFB-induced factor homeobox 2-like, X-linked 2 |
| chr11_+_101875095 | 14.14 |
ENSMUST00000176722.8
ENSMUST00000175972.2 |
Cfap97d1
|
CFAP97 domain containing 1 |
| chr7_+_27222678 | 14.13 |
ENSMUST00000108353.9
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr5_-_143211632 | 13.98 |
ENSMUST00000085733.9
|
Spdye4a
|
speedy/RINGO cell cycle regulator family, member E4A |
| chrX_-_117390434 | 13.87 |
ENSMUST00000073857.6
|
Tgif2lx1
|
TGFB-induced factor homeobox 2-like, X-linked 1 |
| chr3_-_88669551 | 13.73 |
ENSMUST00000183267.2
|
Syt11
|
synaptotagmin XI |
| chr7_+_44146012 | 13.57 |
ENSMUST00000205422.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr12_+_98594388 | 13.57 |
ENSMUST00000048402.12
ENSMUST00000101144.10 ENSMUST00000101146.4 |
Spata7
|
spermatogenesis associated 7 |
| chr7_+_138828391 | 13.51 |
ENSMUST00000093993.5
ENSMUST00000172136.9 |
Pwwp2b
|
PWWP domain containing 2B |
| chr5_-_103359117 | 13.11 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr16_+_20511991 | 12.97 |
ENSMUST00000149543.9
ENSMUST00000232207.2 ENSMUST00000118919.9 |
Fam131a
|
family with sequence similarity 131, member A |
| chr10_+_100428212 | 12.94 |
ENSMUST00000187119.7
ENSMUST00000188736.7 ENSMUST00000191336.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr18_-_3281752 | 12.92 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr15_+_79975520 | 12.91 |
ENSMUST00000009728.13
ENSMUST00000009727.12 |
Syngr1
|
synaptogyrin 1 |
| chr8_-_56359983 | 12.87 |
ENSMUST00000053441.5
|
Adam29
|
a disintegrin and metallopeptidase domain 29 |
| chr3_-_117153802 | 12.83 |
ENSMUST00000197743.2
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr11_+_87457544 | 12.79 |
ENSMUST00000060360.7
|
Septin4
|
septin 4 |
| chr7_-_39062584 | 12.77 |
ENSMUST00000108017.2
|
Gm5114
|
predicted gene 5114 |
| chr7_+_44146029 | 12.75 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr2_-_153712996 | 12.73 |
ENSMUST00000028982.5
|
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr12_-_102709884 | 12.72 |
ENSMUST00000173760.9
ENSMUST00000178384.2 |
Moap1
|
modulator of apoptosis 1 |
| chr7_+_40636967 | 12.70 |
ENSMUST00000206529.2
ENSMUST00000171664.2 |
4930433I11Rik
|
RIKEN cDNA 4930433I11 gene |
| chr16_-_50411484 | 12.65 |
ENSMUST00000062439.6
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr2_-_79738734 | 12.64 |
ENSMUST00000090756.11
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_-_141649003 | 12.56 |
ENSMUST00000039926.10
|
Dusp8
|
dual specificity phosphatase 8 |
| chr18_+_52779281 | 12.55 |
ENSMUST00000118724.8
ENSMUST00000091904.6 |
1700034E13Rik
|
RIKEN cDNA 1700034E13 gene |
| chr1_+_63312420 | 12.53 |
ENSMUST00000239483.2
ENSMUST00000114132.8 ENSMUST00000126932.2 |
Zdbf2
|
zinc finger, DBF-type containing 2 |
| chr1_-_93029547 | 12.38 |
ENSMUST00000112958.9
ENSMUST00000186861.2 ENSMUST00000171556.8 |
Kif1a
|
kinesin family member 1A |
| chr5_+_107645626 | 12.37 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr10_+_53213763 | 12.36 |
ENSMUST00000219491.2
ENSMUST00000163319.9 ENSMUST00000220197.2 ENSMUST00000046221.8 ENSMUST00000218468.2 ENSMUST00000219921.2 |
Pln
|
phospholamban |
| chr4_-_11966367 | 12.34 |
ENSMUST00000056050.5
ENSMUST00000108299.2 ENSMUST00000108297.3 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr11_-_59054107 | 12.26 |
ENSMUST00000069631.3
|
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr11_+_87457479 | 12.24 |
ENSMUST00000239011.2
|
Septin4
|
septin 4 |
| chr2_+_144665576 | 12.15 |
ENSMUST00000028918.4
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr6_-_149090146 | 12.12 |
ENSMUST00000095319.10
ENSMUST00000141346.2 ENSMUST00000111535.8 |
Amn1
|
antagonist of mitotic exit network 1 |
| chr2_+_143388062 | 12.09 |
ENSMUST00000028905.10
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr14_+_66581745 | 11.97 |
ENSMUST00000152093.8
ENSMUST00000074523.13 |
Stmn4
|
stathmin-like 4 |
| chr2_-_120439981 | 11.97 |
ENSMUST00000133612.2
ENSMUST00000102498.8 ENSMUST00000102499.8 |
Lrrc57
|
leucine rich repeat containing 57 |
| chr16_-_92118313 | 11.84 |
ENSMUST00000062638.8
|
Fam243
|
family with sequence similarity 243 |
| chr12_+_86129329 | 11.79 |
ENSMUST00000054565.8
ENSMUST00000222821.2 ENSMUST00000222905.2 |
Ift43
|
intraflagellar transport 43 |
| chr6_+_24528143 | 11.79 |
ENSMUST00000031696.10
|
Asb15
|
ankyrin repeat and SOCS box-containing 15 |
| chr3_+_65573644 | 11.62 |
ENSMUST00000099075.4
ENSMUST00000177434.3 |
Lekr1
|
leucine, glutamate and lysine rich 1 |
| chrX_+_163289318 | 11.60 |
ENSMUST00000033756.3
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr16_-_4698148 | 11.57 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chrX_+_142447361 | 11.56 |
ENSMUST00000126592.8
ENSMUST00000156449.8 ENSMUST00000155215.8 ENSMUST00000112865.8 |
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr4_-_106585127 | 11.54 |
ENSMUST00000106770.8
ENSMUST00000145044.2 |
Mroh7
|
maestro heat-like repeat family member 7 |
| chr2_+_177783713 | 11.45 |
ENSMUST00000103066.10
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr6_+_54658609 | 11.29 |
ENSMUST00000190641.7
ENSMUST00000187701.2 |
Mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr11_-_32774562 | 11.23 |
ENSMUST00000109365.8
ENSMUST00000020508.4 |
Smim23
|
small integral membrane protein 23 |
| chr10_-_53252210 | 11.17 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr12_+_86781154 | 10.94 |
ENSMUST00000095527.6
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr2_+_31985528 | 10.93 |
ENSMUST00000057423.6
ENSMUST00000140762.2 |
Plpp7
|
phospholipid phosphatase 7 (inactive) |
| chr1_-_93029532 | 10.75 |
ENSMUST00000171796.8
|
Kif1a
|
kinesin family member 1A |
| chr17_+_9068805 | 10.73 |
ENSMUST00000115720.8
|
Pde10a
|
phosphodiesterase 10A |
| chr2_+_155907100 | 10.70 |
ENSMUST00000038860.12
|
Spag4
|
sperm associated antigen 4 |
| chr2_+_155224105 | 10.69 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr10_+_59942274 | 10.69 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr2_-_35257741 | 10.68 |
ENSMUST00000028243.2
|
4930568D16Rik
|
RIKEN cDNA 4930568D16 gene |
| chr17_+_79359617 | 10.65 |
ENSMUST00000233916.2
|
Qpct
|
glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
| chr12_+_72488625 | 10.58 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr15_-_103356880 | 10.53 |
ENSMUST00000065978.9
|
Gtsf2
|
gametocyte specific factor 2 |
| chr6_-_70895899 | 10.53 |
ENSMUST00000063456.5
|
Tex37
|
testis expressed 37 |
| chr2_+_128433125 | 10.45 |
ENSMUST00000155430.8
|
Spdye4c
|
speedy/RINGO cell cycle regulator family, member E4C |
| chrX_+_165021919 | 10.34 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr16_-_3725515 | 10.31 |
ENSMUST00000177221.2
ENSMUST00000177323.8 |
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr10_+_59942020 | 10.17 |
ENSMUST00000121820.9
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr6_+_41661356 | 10.15 |
ENSMUST00000031900.6
|
Llcfc1
|
LLLL and CFNLAS motif containing 1 |
| chr17_-_56783376 | 10.13 |
ENSMUST00000223859.2
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr15_-_83989801 | 10.09 |
ENSMUST00000229826.2
ENSMUST00000082365.6 |
Sult4a1
|
sulfotransferase family 4A, member 1 |
| chr8_-_105019806 | 10.08 |
ENSMUST00000212492.2
ENSMUST00000034344.10 |
Cmtm2a
|
CKLF-like MARVEL transmembrane domain containing 2A |
| chr8_+_26339646 | 10.05 |
ENSMUST00000098858.11
|
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr9_-_20638233 | 10.03 |
ENSMUST00000217198.2
|
Olfm2
|
olfactomedin 2 |
| chr12_+_86781141 | 9.99 |
ENSMUST00000223308.2
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr11_-_69791712 | 9.97 |
ENSMUST00000108621.9
ENSMUST00000100969.9 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr4_-_41774097 | 9.89 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr14_-_8457069 | 9.85 |
ENSMUST00000022257.4
|
Atxn7
|
ataxin 7 |
| chr10_+_29087602 | 9.83 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr8_+_23247760 | 9.76 |
ENSMUST00000033941.7
|
Plat
|
plasminogen activator, tissue |
| chr16_+_49519561 | 9.68 |
ENSMUST00000046777.11
ENSMUST00000142682.9 |
Ift57
|
intraflagellar transport 57 |
| chr19_+_8828132 | 9.65 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr17_+_57182472 | 9.56 |
ENSMUST00000025048.7
|
Acsbg3
|
acyl-CoA synthetase bubblegum family member 3 |
| chr17_-_56783462 | 9.53 |
ENSMUST00000067538.6
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr2_-_120439764 | 9.51 |
ENSMUST00000102496.8
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr16_-_57427179 | 9.50 |
ENSMUST00000114371.5
ENSMUST00000232413.2 |
Cmss1
|
cms small ribosomal subunit 1 |
| chr11_-_94412297 | 9.50 |
ENSMUST00000239238.2
|
Mycbpap
|
MYCBP associated protein |
| chr7_+_40682143 | 9.45 |
ENSMUST00000164422.2
|
Gm4884
|
predicted gene 4884 |
| chr19_+_42040681 | 9.43 |
ENSMUST00000164518.4
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.2 | 39.6 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 7.8 | 7.8 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 7.0 | 41.8 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 5.8 | 110.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 5.3 | 16.0 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 4.6 | 36.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 4.2 | 16.6 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 4.0 | 12.1 | GO:0030070 | insulin processing(GO:0030070) |
| 3.9 | 19.7 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 3.6 | 10.7 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 3.4 | 13.7 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 3.2 | 32.1 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 3.1 | 50.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 3.1 | 21.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 3.1 | 15.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 2.8 | 8.5 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 2.7 | 15.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 2.6 | 26.4 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 2.5 | 22.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.4 | 37.7 | GO:0007343 | egg activation(GO:0007343) |
| 2.3 | 9.3 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 2.3 | 25.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 2.3 | 11.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 2.1 | 6.4 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 2.1 | 14.5 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 2.0 | 10.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 2.0 | 29.6 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 2.0 | 9.8 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 1.9 | 5.8 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 1.9 | 15.0 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 1.9 | 13.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.8 | 84.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 1.8 | 7.3 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 1.8 | 16.5 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 1.8 | 7.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.7 | 20.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.7 | 41.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 1.7 | 10.1 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 1.7 | 10.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 1.7 | 5.0 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 1.7 | 16.6 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.6 | 9.9 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 1.6 | 30.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 1.6 | 23.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 1.5 | 26.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.5 | 27.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.5 | 15.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 1.5 | 19.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 1.4 | 12.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.4 | 5.7 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 1.3 | 20.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 1.2 | 23.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 1.2 | 6.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.2 | 7.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.2 | 7.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 1.2 | 23.4 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 1.2 | 12.7 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 1.1 | 18.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.1 | 11.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.1 | 8.8 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 1.1 | 9.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 1.1 | 27.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 1.1 | 3.2 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 1.1 | 7.5 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 1.1 | 5.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 1.1 | 5.3 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 1.0 | 8.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.0 | 5.0 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 1.0 | 6.0 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 1.0 | 8.0 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 1.0 | 7.0 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 1.0 | 6.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.0 | 6.9 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.0 | 2.9 | GO:0021764 | amygdala development(GO:0021764) |
| 0.9 | 165.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.9 | 4.6 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.9 | 3.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.9 | 6.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.9 | 60.8 | GO:0030317 | sperm motility(GO:0030317) |
| 0.9 | 4.3 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.9 | 6.0 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.8 | 7.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.8 | 12.5 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.8 | 10.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.8 | 3.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.8 | 3.3 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.8 | 4.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.8 | 4.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.8 | 11.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.8 | 11.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.8 | 3.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.8 | 4.6 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.8 | 1.5 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.7 | 7.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.7 | 2.8 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.7 | 2.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.7 | 10.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.7 | 4.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.7 | 2.6 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.6 | 4.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.6 | 6.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.6 | 3.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.6 | 14.0 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.6 | 23.8 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.6 | 3.1 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.6 | 2.5 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.6 | 7.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.6 | 4.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.6 | 2.4 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.6 | 2.4 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.6 | 2.4 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.6 | 3.0 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.6 | 3.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.6 | 1.8 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.6 | 12.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.6 | 1.7 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.6 | 13.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.6 | 1.7 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.6 | 17.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.6 | 12.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.5 | 3.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.5 | 10.4 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.5 | 2.2 | GO:0070099 | regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.5 | 37.4 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.5 | 5.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.5 | 5.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 1.6 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.5 | 5.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.5 | 10.3 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.5 | 11.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.5 | 3.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.5 | 5.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.5 | 32.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.5 | 26.9 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.5 | 11.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.5 | 6.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.5 | 4.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.5 | 3.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.5 | 2.9 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.5 | 1.9 | GO:2000864 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.5 | 2.9 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.5 | 8.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.5 | 7.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.5 | 6.1 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.5 | 3.2 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.5 | 11.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.4 | 1.3 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.4 | 8.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.4 | 2.6 | GO:0015819 | lysine transport(GO:0015819) |
| 0.4 | 5.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.4 | 4.6 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.4 | 2.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.4 | 14.6 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.4 | 5.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.4 | 1.6 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.4 | 5.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.4 | 2.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.4 | 12.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.4 | 1.9 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 6.6 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.4 | 4.8 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 2.2 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.4 | 8.7 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.3 | 5.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 3.5 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 7.8 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.3 | 9.7 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.3 | 7.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 13.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 7.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 1.6 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.3 | 1.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.3 | 3.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.3 | 2.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 5.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.3 | 9.7 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.3 | 32.7 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.3 | 4.9 | GO:0043084 | penile erection(GO:0043084) |
| 0.3 | 2.7 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.3 | 10.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 2.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 2.0 | GO:0042148 | meiotic DNA recombinase assembly(GO:0000707) strand invasion(GO:0042148) |
| 0.3 | 0.8 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.3 | 18.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.3 | 13.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.3 | 1.4 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.3 | 2.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 1.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 6.6 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.3 | 8.7 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.3 | 2.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 7.0 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.3 | 9.1 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 10.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.3 | 27.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 3.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 100.2 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.2 | 13.4 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.2 | 2.8 | GO:2000111 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.2 | 13.2 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.2 | 5.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 3.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 2.1 | GO:0086018 | SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 0.2 | 3.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 1.7 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.2 | 3.6 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.2 | 0.6 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.2 | 3.0 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 16.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.2 | 1.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 2.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.2 | 8.4 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.2 | 2.2 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.2 | 11.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 4.3 | GO:0003341 | cilium movement(GO:0003341) |
| 0.2 | 0.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 6.0 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.7 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 4.6 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.4 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 2.0 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.1 | 14.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.3 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 5.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.8 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 0.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.5 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 3.6 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.6 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 2.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 4.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 3.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 3.4 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 4.7 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 3.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.3 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 | 0.4 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 8.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 2.3 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 2.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 1.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 2.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 4.8 | GO:1901880 | negative regulation of protein depolymerization(GO:1901880) |
| 0.1 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 3.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 4.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.7 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 3.9 | GO:0000578 | embryonic axis specification(GO:0000578) |
| 0.1 | 8.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 5.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 12.6 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 0.7 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.1 | 3.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 3.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 4.5 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 3.9 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 1.1 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 1.9 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 2.2 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 15.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 2.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.3 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 2.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 3.0 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 1.7 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.1 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 1.0 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.4 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 1.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 1.9 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 1.6 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.9 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 0.3 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.6 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) female meiosis chromosome separation(GO:0051309) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 3.5 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.0 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 27.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 6.7 | 33.6 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 6.6 | 26.6 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 3.9 | 112.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 3.8 | 11.3 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 3.3 | 52.1 | GO:0044754 | autolysosome(GO:0044754) |
| 2.3 | 11.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 2.3 | 36.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 2.2 | 11.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 1.9 | 21.0 | GO:0002177 | manchette(GO:0002177) |
| 1.8 | 26.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.7 | 5.2 | GO:1990427 | stereocilia tip link(GO:0002140) myosin VII complex(GO:0031477) stereocilia tip-link density(GO:1990427) upper tip-link density(GO:1990435) |
| 1.7 | 5.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.6 | 7.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.5 | 24.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 1.4 | 18.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 1.4 | 6.9 | GO:0044393 | microspike(GO:0044393) |
| 1.3 | 5.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 1.3 | 2.6 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 1.3 | 5.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.2 | 15.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 1.2 | 14.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.2 | 10.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 1.2 | 11.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 1.2 | 24.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 1.1 | 13.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 1.1 | 44.7 | GO:0097546 | ciliary base(GO:0097546) |
| 1.1 | 5.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 1.0 | 30.8 | GO:0000786 | nucleosome(GO:0000786) |
| 1.0 | 7.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.0 | 7.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.0 | 6.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.9 | 7.6 | GO:0031673 | H zone(GO:0031673) |
| 0.9 | 155.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.9 | 13.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.9 | 13.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.8 | 7.6 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.8 | 40.3 | GO:0031430 | M band(GO:0031430) |
| 0.8 | 18.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.8 | 19.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.8 | 24.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.8 | 2.3 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.7 | 20.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.7 | 6.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.7 | 5.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.7 | 6.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.7 | 2.7 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.7 | 17.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.7 | 8.7 | GO:0005858 | axonemal dynein complex(GO:0005858) inner dynein arm(GO:0036156) |
| 0.6 | 3.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.5 | 36.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.5 | 17.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 1.6 | GO:1990879 | CST complex(GO:1990879) |
| 0.5 | 16.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 5.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 16.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.5 | 22.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.5 | 3.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.5 | 4.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.5 | 26.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.5 | 1.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.5 | 4.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.4 | 3.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.4 | 3.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 11.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.4 | 2.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.4 | 23.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.4 | 1.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 1.4 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.3 | 2.8 | GO:0097413 | Lewy body(GO:0097413) |
| 0.3 | 5.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 2.0 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 20.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 5.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 6.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.3 | 2.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 6.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 3.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 4.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 10.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.3 | 8.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 6.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 3.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 2.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.3 | 1.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 4.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 1.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.3 | 3.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 1.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 7.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 1.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 3.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 9.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.2 | 21.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.2 | 11.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 19.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 2.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 2.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 3.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 5.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 2.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 36.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 2.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 2.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 8.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 0.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 2.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 4.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 3.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 3.1 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 18.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 2.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 4.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 34.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 14.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 2.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 11.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 8.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 2.9 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 5.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 13.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 4.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 9.3 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 31.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 3.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 6.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 13.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 11.1 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 2.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 9.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 2.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.3 | GO:0005655 | ribonuclease MRP complex(GO:0000172) nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 2.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 3.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 2.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 7.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 25.3 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 6.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 4.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 2.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 73.9 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 5.3 | 16.0 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 5.2 | 57.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 5.1 | 15.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 4.6 | 32.1 | GO:0045174 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 3.9 | 19.7 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 3.2 | 12.9 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 2.9 | 8.8 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 2.8 | 16.6 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 2.7 | 24.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 2.6 | 13.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.6 | 20.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 2.5 | 27.6 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 2.4 | 14.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 2.3 | 41.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 2.3 | 6.8 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 2.1 | 8.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 2.1 | 12.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 2.0 | 22.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 2.0 | 19.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.8 | 16.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.8 | 10.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 1.7 | 19.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.7 | 44.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 1.7 | 6.8 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 1.7 | 11.8 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.6 | 19.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 1.6 | 4.8 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 1.6 | 23.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.6 | 28.5 | GO:0043495 | protein anchor(GO:0043495) |
| 1.5 | 10.7 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 1.5 | 8.8 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 1.5 | 7.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 1.4 | 5.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.4 | 12.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.3 | 25.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 1.3 | 10.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.2 | 7.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.2 | 3.5 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 1.1 | 6.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.1 | 4.3 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 1.1 | 60.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 1.0 | 8.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 1.0 | 3.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 1.0 | 9.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 1.0 | 4.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 1.0 | 2.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.0 | 11.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.9 | 8.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.9 | 7.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.9 | 51.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.9 | 6.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.8 | 7.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.8 | 8.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.8 | 7.5 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.8 | 3.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.8 | 18.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.8 | 60.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.7 | 5.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.7 | 17.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.7 | 4.5 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.7 | 15.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.7 | 2.8 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.7 | 26.9 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.7 | 17.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.7 | 6.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 3.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.7 | 13.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.6 | 8.2 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.6 | 2.5 | GO:0004079 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.6 | 5.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.6 | 6.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.6 | 20.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.6 | 1.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.6 | 3.4 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.6 | 36.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.6 | 31.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.5 | 10.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.5 | 3.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 12.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.5 | 2.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.5 | 3.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 25.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.5 | 1.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 1.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.5 | 19.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.5 | 51.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.5 | 4.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 6.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.5 | 1.4 | GO:0032139 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.5 | 2.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 2.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 2.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 3.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.4 | 10.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 122.6 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.3 | 2.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.3 | 2.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 12.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.3 | 8.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 23.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 14.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 2.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 7.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 6.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.3 | 1.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.3 | 6.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 3.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 8.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 2.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 10.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 2.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 1.0 | GO:0051381 | histamine binding(GO:0051381) |
| 0.3 | 63.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.3 | 4.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 1.5 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.3 | 4.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 7.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 5.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 4.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 3.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 7.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) hexose transmembrane transporter activity(GO:0015149) |
| 0.2 | 1.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 4.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 5.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 8.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 7.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 11.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 5.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 17.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 6.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 7.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 5.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 3.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 4.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 1.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 3.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 2.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 6.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 14.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 2.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 2.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 0.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.3 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 2.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 31.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 4.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 6.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.7 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 8.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 2.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.4 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 15.3 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 11.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 14.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 6.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.5 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 0.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 1.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 8.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 16.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 2.1 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 2.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 5.8 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 2.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 2.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 1.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 2.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 18.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 2.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 3.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 16.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 2.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 7.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 19.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.8 | 34.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.6 | 32.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.6 | 31.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.5 | 5.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 27.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.5 | 7.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 9.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 10.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 6.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 4.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 7.3 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 7.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 7.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 6.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 5.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 4.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 2.4 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 1.1 | 37.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 1.0 | 19.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 1.0 | 12.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.8 | 18.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.8 | 20.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.8 | 14.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.8 | 12.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.7 | 2.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.7 | 10.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.7 | 9.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.6 | 29.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.6 | 16.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 13.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.5 | 34.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.5 | 24.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 10.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.5 | 2.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.5 | 4.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 10.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 15.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.4 | 7.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.4 | 7.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.4 | 7.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 6.9 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.3 | 18.7 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.3 | 7.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 5.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 5.6 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.3 | 3.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 3.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 13.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 9.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 3.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 6.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 4.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 15.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 3.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 1.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 3.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 4.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 6.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 7.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 5.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 9.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 4.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 9.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 20.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 4.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.2 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 6.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.5 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |