Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
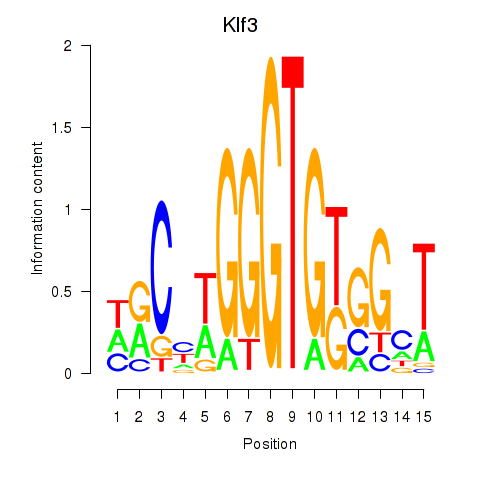
Results for Klf3
Z-value: 0.43
Transcription factors associated with Klf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf3
|
ENSMUSG00000029178.15 | Klf3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf3 | mm39_v1_chr5_+_64960705_64960731 | -0.13 | 2.7e-01 | Click! |
Activity profile of Klf3 motif
Sorted Z-values of Klf3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_82933383 | 3.53 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr9_-_50657800 | 2.72 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr13_+_89687915 | 1.88 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr2_+_163389068 | 1.85 |
ENSMUST00000109411.8
ENSMUST00000018094.13 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr19_+_10018914 | 1.04 |
ENSMUST00000115995.4
|
Fads3
|
fatty acid desaturase 3 |
| chr7_-_140676596 | 0.90 |
ENSMUST00000209199.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr11_+_68858942 | 0.86 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr3_-_108306267 | 0.84 |
ENSMUST00000147251.2
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr9_-_108183356 | 0.84 |
ENSMUST00000192886.6
|
Tcta
|
T cell leukemia translocation altered gene |
| chr17_-_23941757 | 0.84 |
ENSMUST00000178006.8
|
1520401A03Rik
|
RIKEN cDNA 1520401A03 gene |
| chr9_-_81515865 | 0.81 |
ENSMUST00000183482.2
|
Htr1b
|
5-hydroxytryptamine (serotonin) receptor 1B |
| chr9_-_108183140 | 0.76 |
ENSMUST00000195615.2
|
Tcta
|
T cell leukemia translocation altered gene |
| chr19_+_10019023 | 0.75 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr14_+_63284438 | 0.71 |
ENSMUST00000067990.8
ENSMUST00000111203.2 |
Defb42
|
defensin beta 42 |
| chr15_-_36792649 | 0.71 |
ENSMUST00000126184.2
|
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr7_-_140676623 | 0.69 |
ENSMUST00000209352.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr17_+_23883974 | 0.64 |
ENSMUST00000233541.2
|
Bicdl2
|
BICD family like cargo adaptor 2 |
| chr9_-_95697441 | 0.63 |
ENSMUST00000119760.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr10_+_93324624 | 0.58 |
ENSMUST00000129421.8
|
Hal
|
histidine ammonia lyase |
| chr4_-_41774097 | 0.52 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr12_+_87073338 | 0.49 |
ENSMUST00000110187.8
ENSMUST00000156162.8 |
Tmem63c
|
transmembrane protein 63c |
| chr9_-_108183162 | 0.48 |
ENSMUST00000044725.9
|
Tcta
|
T cell leukemia translocation altered gene |
| chr7_-_105218472 | 0.48 |
ENSMUST00000187683.7
ENSMUST00000210079.2 ENSMUST00000187051.7 ENSMUST00000189265.7 ENSMUST00000190369.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr12_+_87073060 | 0.47 |
ENSMUST00000146292.8
|
Tmem63c
|
transmembrane protein 63c |
| chr7_-_105217851 | 0.47 |
ENSMUST00000188368.7
ENSMUST00000187057.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chrX_-_101295787 | 0.42 |
ENSMUST00000050551.10
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr11_+_98592089 | 0.41 |
ENSMUST00000038886.3
|
Csf3
|
colony stimulating factor 3 (granulocyte) |
| chrX_-_101295458 | 0.38 |
ENSMUST00000101336.10
ENSMUST00000136277.2 |
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr1_+_135075377 | 0.35 |
ENSMUST00000125774.2
|
Arl8a
|
ADP-ribosylation factor-like 8A |
| chrX_+_7750558 | 0.35 |
ENSMUST00000208640.2
ENSMUST00000207114.2 ENSMUST00000208633.2 ENSMUST00000208397.2 ENSMUST00000153620.3 ENSMUST00000123277.8 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr3_-_88241735 | 0.33 |
ENSMUST00000107552.2
|
Tmem79
|
transmembrane protein 79 |
| chr2_-_155849839 | 0.33 |
ENSMUST00000086145.10
ENSMUST00000144686.8 ENSMUST00000140657.8 |
6430550D23Rik
|
RIKEN cDNA 6430550D23 gene |
| chr9_-_44624496 | 0.26 |
ENSMUST00000144251.8
ENSMUST00000156918.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr13_+_55468313 | 0.23 |
ENSMUST00000021942.8
|
Prelid1
|
PRELI domain containing 1 |
| chr3_+_89987749 | 0.20 |
ENSMUST00000127955.2
|
Tpm3
|
tropomyosin 3, gamma |
| chr3_+_89173859 | 0.18 |
ENSMUST00000040824.2
|
Dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr7_-_142628298 | 0.11 |
ENSMUST00000148715.8
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr7_-_104246386 | 0.06 |
ENSMUST00000057385.5
|
Olfr655
|
olfactory receptor 655 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.6 | 1.8 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.3 | 1.6 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.2 | 0.8 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 0.8 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.2 | 2.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.2 | 0.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.9 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.8 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 0.7 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.6 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.1 | 0.5 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 | 0.3 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.3 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.2 | GO:1901857 | negative regulation of mitochondrial membrane potential(GO:0010917) positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.9 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.1 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 1.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.8 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.8 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 3.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.6 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |