Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
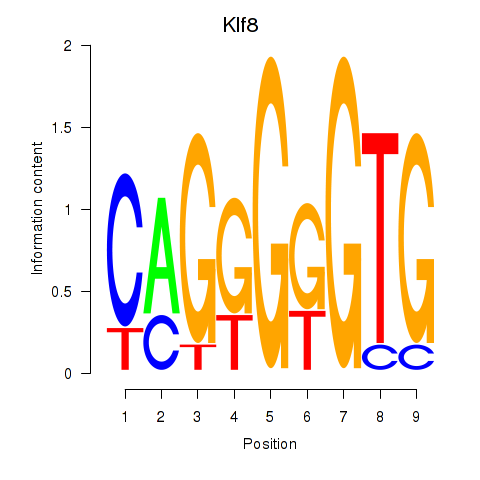
Results for Klf8
Z-value: 0.72
Transcription factors associated with Klf8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf8
|
ENSMUSG00000041649.14 | Klf8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf8 | mm39_v1_chrX_+_152142571_152142609 | -0.52 | 2.4e-06 | Click! |
Activity profile of Klf8 motif
Sorted Z-values of Klf8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_31079177 | 4.71 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chrX_-_84820209 | 4.51 |
ENSMUST00000142152.2
ENSMUST00000156390.8 |
Gk
|
glycerol kinase |
| chrX_-_84820250 | 3.82 |
ENSMUST00000113978.9
|
Gk
|
glycerol kinase |
| chr10_+_75399920 | 3.64 |
ENSMUST00000141062.8
ENSMUST00000152657.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr11_+_66847446 | 3.38 |
ENSMUST00000211300.2
ENSMUST00000150220.2 |
Tmem238l
|
transmembrane protein 238 like |
| chr18_+_61058684 | 3.37 |
ENSMUST00000102888.10
ENSMUST00000025519.11 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr13_-_73848807 | 3.30 |
ENSMUST00000022048.6
|
Slc6a19
|
solute carrier family 6 (neurotransmitter transporter), member 19 |
| chr7_-_30623592 | 3.26 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr5_-_90788323 | 3.23 |
ENSMUST00000202784.4
ENSMUST00000031317.10 ENSMUST00000201370.2 |
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_+_94284739 | 3.01 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr6_-_131293361 | 2.96 |
ENSMUST00000121078.2
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr18_+_61058716 | 2.88 |
ENSMUST00000115297.8
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr11_+_102036356 | 2.81 |
ENSMUST00000055409.6
|
Nags
|
N-acetylglutamate synthase |
| chr1_+_135746330 | 2.66 |
ENSMUST00000038760.10
|
Lad1
|
ladinin |
| chr3_+_94284812 | 2.58 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr10_+_127595639 | 2.48 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr13_+_120151982 | 2.47 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr5_-_90788460 | 2.44 |
ENSMUST00000202704.4
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_+_31078911 | 2.27 |
ENSMUST00000201571.4
|
Khk
|
ketohexokinase |
| chr15_+_78290896 | 2.18 |
ENSMUST00000167140.8
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr2_+_143757193 | 2.10 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr9_-_44231526 | 2.04 |
ENSMUST00000214602.2
ENSMUST00000065080.10 |
C2cd2l
|
C2 calcium-dependent domain containing 2-like |
| chr15_+_78290975 | 1.97 |
ENSMUST00000043865.8
ENSMUST00000231159.2 ENSMUST00000169133.8 |
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr7_-_28947882 | 1.95 |
ENSMUST00000032808.6
|
2200002D01Rik
|
RIKEN cDNA 2200002D01 gene |
| chr17_-_36395115 | 1.88 |
ENSMUST00000184502.8
ENSMUST00000183560.8 ENSMUST00000183999.8 |
H2-Bl
|
histocompatibility 2, blastocyst |
| chr9_-_34967081 | 1.88 |
ENSMUST00000215463.2
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr1_+_187340952 | 1.84 |
ENSMUST00000127489.8
|
Esrrg
|
estrogen-related receptor gamma |
| chrX_-_50294652 | 1.79 |
ENSMUST00000114875.8
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr8_-_105991219 | 1.77 |
ENSMUST00000034359.10
|
Tradd
|
TNFRSF1A-associated via death domain |
| chr15_-_89258012 | 1.73 |
ENSMUST00000167643.4
|
Sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr3_-_121608809 | 1.73 |
ENSMUST00000197383.5
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr8_-_71112295 | 1.70 |
ENSMUST00000211715.2
ENSMUST00000210307.2 ENSMUST00000209768.2 ENSMUST00000070173.9 |
Pgpep1
|
pyroglutamyl-peptidase I |
| chr5_-_123270702 | 1.69 |
ENSMUST00000031401.6
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr5_-_123270449 | 1.68 |
ENSMUST00000186469.7
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr12_+_28725218 | 1.66 |
ENSMUST00000020957.13
|
Adi1
|
acireductone dioxygenase 1 |
| chr12_-_114286421 | 1.65 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8 |
| chr6_+_124908439 | 1.63 |
ENSMUST00000032214.14
|
Mlf2
|
myeloid leukemia factor 2 |
| chr6_-_52185674 | 1.60 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr7_-_127529238 | 1.57 |
ENSMUST00000032988.10
ENSMUST00000206124.2 |
Prss8
|
protease, serine 8 (prostasin) |
| chr11_+_72889889 | 1.55 |
ENSMUST00000021141.14
|
P2rx1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr6_+_124908389 | 1.52 |
ENSMUST00000180095.4
|
Mlf2
|
myeloid leukemia factor 2 |
| chr3_-_121608859 | 1.51 |
ENSMUST00000029770.8
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr17_-_34219225 | 1.50 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr17_-_25179635 | 1.49 |
ENSMUST00000024981.9
|
Jpt2
|
Jupiter microtubule associated homolog 2 |
| chrX_+_74460275 | 1.49 |
ENSMUST00000118428.8
ENSMUST00000114074.8 ENSMUST00000133781.8 |
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr15_-_89258034 | 1.46 |
ENSMUST00000228977.2
|
Sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr7_-_28246530 | 1.41 |
ENSMUST00000239002.2
ENSMUST00000057974.4 |
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr7_+_51528715 | 1.36 |
ENSMUST00000051912.13
|
Gas2
|
growth arrest specific 2 |
| chr7_-_126625657 | 1.34 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr13_-_100037149 | 1.34 |
ENSMUST00000022150.8
|
Cartpt
|
CART prepropeptide |
| chr11_-_35871300 | 1.29 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr1_-_135846937 | 1.27 |
ENSMUST00000027667.13
|
Pkp1
|
plakophilin 1 |
| chr11_+_83637766 | 1.27 |
ENSMUST00000070832.3
|
Wfdc21
|
WAP four-disulfide core domain 21 |
| chrX_-_72965524 | 1.26 |
ENSMUST00000114389.10
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chrX_+_74460234 | 1.22 |
ENSMUST00000033544.14
|
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr10_-_90959853 | 1.20 |
ENSMUST00000170810.8
ENSMUST00000076694.13 |
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chrX_+_16485937 | 1.19 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr10_-_90959817 | 1.19 |
ENSMUST00000164505.2
|
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr17_-_46956920 | 1.18 |
ENSMUST00000233974.2
|
Klc4
|
kinesin light chain 4 |
| chrX_-_72965434 | 1.18 |
ENSMUST00000096316.4
ENSMUST00000114390.8 ENSMUST00000114391.10 ENSMUST00000114387.8 |
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr1_+_172327569 | 1.15 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr14_-_55828511 | 1.12 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr6_+_124908341 | 1.12 |
ENSMUST00000203021.3
|
Mlf2
|
myeloid leukemia factor 2 |
| chr6_+_41520150 | 1.12 |
ENSMUST00000103295.2
|
Trbj2-3
|
T cell receptor beta joining 2-3 |
| chr19_-_6117815 | 1.12 |
ENSMUST00000162575.8
ENSMUST00000159084.8 ENSMUST00000161718.8 ENSMUST00000162810.8 ENSMUST00000025713.12 ENSMUST00000113543.9 ENSMUST00000160417.8 ENSMUST00000161528.2 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr7_+_64151838 | 1.11 |
ENSMUST00000205604.2
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr14_+_122771734 | 1.10 |
ENSMUST00000154206.8
ENSMUST00000038374.13 ENSMUST00000135578.8 |
Pcca
|
propionyl-Coenzyme A carboxylase, alpha polypeptide |
| chr9_+_43655230 | 1.10 |
ENSMUST00000034510.9
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr2_+_172187485 | 1.09 |
ENSMUST00000028995.5
|
Fam210b
|
family with sequence similarity 210, member B |
| chr7_+_51528788 | 1.06 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr14_-_30645503 | 1.05 |
ENSMUST00000227995.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr5_+_115373895 | 1.02 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr19_-_29339224 | 1.01 |
ENSMUST00000126800.8
ENSMUST00000152936.8 ENSMUST00000143467.8 ENSMUST00000016639.12 |
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr19_+_8641369 | 0.99 |
ENSMUST00000035444.10
ENSMUST00000163785.2 |
Chrm1
|
cholinergic receptor, muscarinic 1, CNS |
| chrX_-_72965536 | 0.99 |
ENSMUST00000033763.15
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr4_-_130068902 | 0.97 |
ENSMUST00000105998.8
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr3_+_129007599 | 0.97 |
ENSMUST00000042587.12
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr17_-_26087696 | 0.97 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr1_+_135945798 | 0.96 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr14_-_47025724 | 0.96 |
ENSMUST00000146629.3
ENSMUST00000015903.12 |
Cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr8_+_105991280 | 0.96 |
ENSMUST00000036221.12
|
Fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr1_+_135945705 | 0.94 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr11_-_76462353 | 0.94 |
ENSMUST00000072740.13
|
Abr
|
active BCR-related gene |
| chr15_+_5173342 | 0.92 |
ENSMUST00000051186.9
ENSMUST00000228218.2 |
Prkaa1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chrM_+_14138 | 0.91 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrX_+_56008685 | 0.90 |
ENSMUST00000096431.10
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr15_-_98507913 | 0.90 |
ENSMUST00000226500.2
ENSMUST00000227501.2 |
Adcy6
|
adenylate cyclase 6 |
| chr6_+_41519654 | 0.89 |
ENSMUST00000103293.2
|
Trbj2-1
|
T cell receptor beta joining 2-1 |
| chr17_-_24915121 | 0.89 |
ENSMUST00000046839.10
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr7_-_126625617 | 0.88 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr4_-_150998857 | 0.87 |
ENSMUST00000105675.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr11_-_5900019 | 0.87 |
ENSMUST00000102920.4
|
Gck
|
glucokinase |
| chr10_+_62088104 | 0.86 |
ENSMUST00000020278.6
|
Tacr2
|
tachykinin receptor 2 |
| chr11_-_69838971 | 0.86 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr2_-_65068917 | 0.86 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr19_-_6899121 | 0.85 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr2_-_90735171 | 0.84 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr1_-_184615415 | 0.84 |
ENSMUST00000048308.6
|
C130074G19Rik
|
RIKEN cDNA C130074G19 gene |
| chr17_-_26014613 | 0.83 |
ENSMUST00000235889.2
|
Gm50367
|
predicted gene, 50367 |
| chr18_+_36414122 | 0.82 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr19_+_6547790 | 0.81 |
ENSMUST00000113458.8
ENSMUST00000113459.2 |
Nrxn2
|
neurexin II |
| chr2_+_85551751 | 0.79 |
ENSMUST00000055517.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr11_-_86574586 | 0.78 |
ENSMUST00000018315.10
|
Vmp1
|
vacuole membrane protein 1 |
| chr12_-_113236868 | 0.78 |
ENSMUST00000223335.2
ENSMUST00000137336.3 |
Ighe
|
Immunoglobulin heavy constant epsilon |
| chr11_-_102837514 | 0.77 |
ENSMUST00000057849.6
|
C1ql1
|
complement component 1, q subcomponent-like 1 |
| chr5_-_108808649 | 0.77 |
ENSMUST00000053913.13
|
Dgkq
|
diacylglycerol kinase, theta |
| chr19_-_6899173 | 0.76 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr6_+_41520287 | 0.75 |
ENSMUST00000103296.2
|
Trbj2-4
|
T cell receptor beta joining 2-4 |
| chr1_+_172327812 | 0.75 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr16_-_94657531 | 0.74 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr12_+_105302853 | 0.72 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr5_+_124045552 | 0.72 |
ENSMUST00000166233.2
|
Denr
|
density-regulated protein |
| chr3_+_156267772 | 0.71 |
ENSMUST00000175773.2
|
Negr1
|
neuronal growth regulator 1 |
| chr11_-_115310743 | 0.71 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chrM_-_14061 | 0.71 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr12_-_103597663 | 0.70 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr9_+_37466989 | 0.70 |
ENSMUST00000213126.2
|
Siae
|
sialic acid acetylesterase |
| chr9_-_114610879 | 0.70 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr5_+_130477642 | 0.70 |
ENSMUST00000111288.4
|
Caln1
|
calneuron 1 |
| chr17_-_45912811 | 0.69 |
ENSMUST00000169137.3
ENSMUST00000208801.2 |
Mymx
|
myomixer, myoblast fusion factor |
| chr5_+_147206769 | 0.69 |
ENSMUST00000085591.7
|
Pdx1
|
pancreatic and duodenal homeobox 1 |
| chr14_-_70864448 | 0.69 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr1_-_106641940 | 0.69 |
ENSMUST00000112751.2
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chrX_-_59449137 | 0.69 |
ENSMUST00000033480.13
ENSMUST00000101527.3 |
Atp11c
|
ATPase, class VI, type 11C |
| chr9_-_106035308 | 0.68 |
ENSMUST00000159809.2
ENSMUST00000162562.2 ENSMUST00000036382.13 |
Glyctk
|
glycerate kinase |
| chr1_-_66902429 | 0.68 |
ENSMUST00000027153.6
|
Acadl
|
acyl-Coenzyme A dehydrogenase, long-chain |
| chr7_-_27055405 | 0.67 |
ENSMUST00000003857.7
|
Shkbp1
|
Sh3kbp1 binding protein 1 |
| chr2_-_26096547 | 0.67 |
ENSMUST00000028302.8
|
Lhx3
|
LIM homeobox protein 3 |
| chr5_+_129924564 | 0.66 |
ENSMUST00000041466.14
|
Zbed5
|
zinc finger, BED type containing 5 |
| chr8_+_86567600 | 0.66 |
ENSMUST00000053771.14
ENSMUST00000161850.8 |
Phkb
|
phosphorylase kinase beta |
| chr1_-_10038030 | 0.65 |
ENSMUST00000185184.2
|
Tcf24
|
transcription factor 24 |
| chr9_+_108765701 | 0.64 |
ENSMUST00000026743.14
ENSMUST00000194047.3 |
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr14_-_30850795 | 0.63 |
ENSMUST00000049732.11
ENSMUST00000090205.5 ENSMUST00000064032.10 |
Smim4
|
small integral membrane protein 4 |
| chr12_-_118265163 | 0.63 |
ENSMUST00000221844.2
|
Sp4
|
trans-acting transcription factor 4 |
| chr12_-_104831335 | 0.63 |
ENSMUST00000109936.3
|
Clmn
|
calmin |
| chr6_+_41519884 | 0.62 |
ENSMUST00000103294.2
|
Trbj2-2
|
T cell receptor beta joining 2-2 |
| chr7_+_64151435 | 0.62 |
ENSMUST00000032732.15
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr10_+_127595590 | 0.62 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr19_+_5540591 | 0.61 |
ENSMUST00000237122.2
|
Cfl1
|
cofilin 1, non-muscle |
| chr2_-_85193402 | 0.61 |
ENSMUST00000111597.3
|
Olfr988
|
olfactory receptor 988 |
| chr10_+_43355113 | 0.61 |
ENSMUST00000040147.8
|
Bend3
|
BEN domain containing 3 |
| chr11_+_11634967 | 0.61 |
ENSMUST00000141436.8
ENSMUST00000126058.8 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr2_+_27599259 | 0.58 |
ENSMUST00000100251.9
|
Rxra
|
retinoid X receptor alpha |
| chr1_+_125488747 | 0.57 |
ENSMUST00000027580.11
|
Slc35f5
|
solute carrier family 35, member F5 |
| chr5_+_88712840 | 0.57 |
ENSMUST00000196894.5
ENSMUST00000198965.5 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr7_+_101070897 | 0.57 |
ENSMUST00000163751.10
ENSMUST00000211368.2 ENSMUST00000166652.2 |
Pde2a
|
phosphodiesterase 2A, cGMP-stimulated |
| chr2_-_52566583 | 0.56 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr9_-_106035332 | 0.56 |
ENSMUST00000112543.9
|
Glyctk
|
glycerate kinase |
| chr19_+_6952580 | 0.56 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr10_+_23846604 | 0.56 |
ENSMUST00000092659.4
|
Taar5
|
trace amine-associated receptor 5 |
| chr19_+_5540483 | 0.56 |
ENSMUST00000209469.2
ENSMUST00000116560.3 |
Cfl1
|
cofilin 1, non-muscle |
| chr8_+_63404395 | 0.56 |
ENSMUST00000119068.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr17_-_47015928 | 0.56 |
ENSMUST00000002839.9
ENSMUST00000233988.2 |
Ppp2r5d
|
protein phosphatase 2, regulatory subunit B', delta |
| chr2_-_28730286 | 0.55 |
ENSMUST00000037117.6
ENSMUST00000171404.8 |
Gtf3c4
|
general transcription factor IIIC, polypeptide 4 |
| chr4_+_108736350 | 0.54 |
ENSMUST00000106651.9
|
Rab3b
|
RAB3B, member RAS oncogene family |
| chrX_+_7750483 | 0.54 |
ENSMUST00000115663.10
ENSMUST00000096514.11 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chrX_-_166906307 | 0.54 |
ENSMUST00000112149.9
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr5_+_31205971 | 0.53 |
ENSMUST00000013766.13
ENSMUST00000201773.4 ENSMUST00000200748.4 ENSMUST00000201136.2 |
Atraid
|
all-trans retinoic acid induced differentiation factor |
| chr10_-_5755412 | 0.53 |
ENSMUST00000019907.8
|
Fbxo5
|
F-box protein 5 |
| chr4_+_155896946 | 0.53 |
ENSMUST00000030944.11
|
Ccnl2
|
cyclin L2 |
| chr18_+_84738144 | 0.52 |
ENSMUST00000161429.3
ENSMUST00000052501.8 |
Dipk1c
|
divergent protein kinase domain 1C |
| chr11_+_114689750 | 0.52 |
ENSMUST00000045319.9
ENSMUST00000106584.2 |
Gpr142
|
G protein-coupled receptor 142 |
| chr11_-_83193412 | 0.51 |
ENSMUST00000176374.2
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr1_+_158190090 | 0.51 |
ENSMUST00000194369.6
ENSMUST00000195311.6 |
Astn1
|
astrotactin 1 |
| chr14_-_68170873 | 0.51 |
ENSMUST00000039135.6
|
Dock5
|
dedicator of cytokinesis 5 |
| chr17_+_25992761 | 0.50 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr9_-_31824758 | 0.50 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chr17_+_46957151 | 0.50 |
ENSMUST00000002844.14
ENSMUST00000113429.8 ENSMUST00000113430.2 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr7_-_139953579 | 0.49 |
ENSMUST00000074177.3
|
Olfr530
|
olfactory receptor 530 |
| chr12_+_109418759 | 0.49 |
ENSMUST00000056110.15
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr18_-_42712717 | 0.49 |
ENSMUST00000054738.5
|
Gpr151
|
G protein-coupled receptor 151 |
| chr17_-_45912993 | 0.49 |
ENSMUST00000178858.3
|
Mymx
|
myomixer, myoblast fusion factor |
| chr3_-_92441809 | 0.48 |
ENSMUST00000193521.2
|
2310046K23Rik
|
RIKEN cDNA 2310046K23 gene |
| chr5_+_139408906 | 0.48 |
ENSMUST00000066211.5
|
Gper1
|
G protein-coupled estrogen receptor 1 |
| chr13_+_38010879 | 0.48 |
ENSMUST00000149745.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr19_+_6952319 | 0.47 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr10_+_43354807 | 0.47 |
ENSMUST00000167488.9
|
Bend3
|
BEN domain containing 3 |
| chrX_+_161543423 | 0.46 |
ENSMUST00000112326.8
|
Rbbp7
|
retinoblastoma binding protein 7, chromatin remodeling factor |
| chr15_+_79999643 | 0.46 |
ENSMUST00000135727.2
|
Syngr1
|
synaptogyrin 1 |
| chr8_-_85414220 | 0.46 |
ENSMUST00000238449.2
ENSMUST00000238687.2 |
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr10_-_81463091 | 0.45 |
ENSMUST00000143424.2
ENSMUST00000119324.8 |
Sirt6
|
sirtuin 6 |
| chr13_+_55517545 | 0.45 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr12_-_69274936 | 0.45 |
ENSMUST00000221411.2
ENSMUST00000021359.7 |
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr7_+_112026712 | 0.44 |
ENSMUST00000106643.8
ENSMUST00000033030.14 |
Parva
|
parvin, alpha |
| chr11_-_76386190 | 0.44 |
ENSMUST00000108408.9
|
Abr
|
active BCR-related gene |
| chr11_-_99176086 | 0.44 |
ENSMUST00000017255.4
|
Krt24
|
keratin 24 |
| chr14_-_30850881 | 0.44 |
ENSMUST00000203261.3
|
Smim4
|
small integral membrane protein 4 |
| chr2_-_168583670 | 0.43 |
ENSMUST00000029060.11
|
Atp9a
|
ATPase, class II, type 9A |
| chr8_+_114932312 | 0.43 |
ENSMUST00000049509.7
ENSMUST00000150963.2 |
Vat1l
|
vesicle amine transport protein 1 like |
| chrX_+_73348598 | 0.43 |
ENSMUST00000015435.11
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr3_+_156267429 | 0.42 |
ENSMUST00000074015.11
|
Negr1
|
neuronal growth regulator 1 |
| chr11_+_60428788 | 0.42 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr10_+_79716876 | 0.42 |
ENSMUST00000166201.2
|
Prtn3
|
proteinase 3 |
| chr4_+_108736260 | 0.42 |
ENSMUST00000106650.9
|
Rab3b
|
RAB3B, member RAS oncogene family |
| chr15_-_37792237 | 0.41 |
ENSMUST00000168992.8
ENSMUST00000148652.9 |
Ncald
|
neurocalcin delta |
| chr9_-_96513529 | 0.41 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr8_+_22682816 | 0.40 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr2_+_156681991 | 0.40 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chrX_+_161543384 | 0.39 |
ENSMUST00000033720.12
ENSMUST00000112327.8 |
Rbbp7
|
retinoblastoma binding protein 7, chromatin remodeling factor |
| chr19_-_53026965 | 0.39 |
ENSMUST00000183274.8
ENSMUST00000182097.2 |
Xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 7.0 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 1.7 | 8.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.8 | 3.3 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.8 | 3.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.7 | 2.7 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.5 | 3.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.5 | 5.6 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.5 | 3.6 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.4 | 6.3 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.4 | 3.4 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.4 | 4.1 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.4 | 1.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.4 | 2.8 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.3 | 1.0 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.3 | 1.6 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.3 | 0.9 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.3 | 0.9 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.3 | 1.5 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.3 | 1.3 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.3 | 1.6 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.2 | 0.7 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.9 | GO:1903189 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.2 | 0.9 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.2 | 0.6 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.2 | 2.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 1.0 | GO:0021763 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.2 | 1.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 0.5 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.2 | 0.7 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.2 | 1.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 | 0.9 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.2 | 0.7 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) |
| 0.2 | 1.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 0.7 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.2 | 0.7 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 0.5 | GO:0051385 | response to mineralocorticoid(GO:0051385) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.2 | 0.8 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 0.8 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 3.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.6 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 1.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.9 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 0.7 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.4 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.2 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.6 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 0.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 1.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.6 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 1.0 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.5 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.2 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 2.5 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.4 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 0.9 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 0.2 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 1.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 1.6 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.1 | 3.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.4 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.2 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.1 | 0.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.5 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.4 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 3.0 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 0.3 | GO:0032423 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.6 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.5 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.3 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.5 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 2.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.7 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.4 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.6 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.4 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.3 | GO:0060455 | gastric emptying(GO:0035483) response to immobilization stress(GO:0035902) negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 1.5 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.3 | GO:0043585 | nose morphogenesis(GO:0043585) alveolar primary septum development(GO:0061143) |
| 0.0 | 1.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.6 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 1.1 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.7 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.6 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.0 | 0.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 2.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.5 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 1.1 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.7 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0002362 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.7 | 2.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.3 | 1.6 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.3 | 3.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 0.7 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.2 | 0.6 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.2 | 0.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 1.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 0.8 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.5 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.5 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 3.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.7 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 1.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 10.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0044279 | growing cell tip(GO:0035838) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 2.4 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.0 | 0.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 11.5 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 2.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 1.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 1.7 | 8.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 1.4 | 4.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.9 | 5.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.8 | 2.5 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.5 | 2.8 | GO:0034618 | arginine binding(GO:0034618) |
| 0.4 | 3.4 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.4 | 6.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.4 | 1.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.4 | 3.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 1.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 3.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.3 | 1.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.3 | 1.9 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 0.7 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.2 | 0.2 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.2 | 0.9 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.2 | 0.9 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 2.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 0.8 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.2 | 0.6 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.2 | 0.9 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 0.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 0.7 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 0.5 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 1.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 2.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 2.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.9 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 3.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.4 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.9 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.1 | 1.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 1.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.4 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.3 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.2 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.5 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 3.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0044547 | rRNA primary transcript binding(GO:0042134) DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 2.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.1 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 1.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 2.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 4.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 4.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 1.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 3.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 8.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 1.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 9.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.7 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |