Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Lef1
Z-value: 1.33
Transcription factors associated with Lef1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lef1
|
ENSMUSG00000027985.15 | Lef1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lef1 | mm39_v1_chr3_+_130904000_130904120 | -0.34 | 3.7e-03 | Click! |
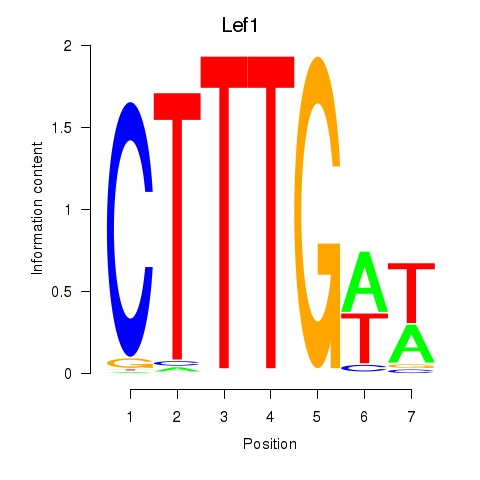
Activity profile of Lef1 motif
Sorted Z-values of Lef1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Lef1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_76842263 | 12.58 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr15_-_58078274 | 8.23 |
ENSMUST00000022986.8
|
Fbxo32
|
F-box protein 32 |
| chr2_+_121188195 | 7.37 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr14_+_102078038 | 7.16 |
ENSMUST00000159314.8
|
Lmo7
|
LIM domain only 7 |
| chr11_+_96162283 | 7.14 |
ENSMUST00000000010.9
ENSMUST00000174042.3 |
Hoxb9
|
homeobox B9 |
| chr2_-_60503998 | 6.93 |
ENSMUST00000059888.15
ENSMUST00000154764.2 |
Itgb6
|
integrin beta 6 |
| chr5_-_134975773 | 6.88 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr16_-_97412169 | 6.61 |
ENSMUST00000232141.2
ENSMUST00000000395.8 |
Tmprss2
|
transmembrane protease, serine 2 |
| chr16_+_96001865 | 6.39 |
ENSMUST00000171181.9
ENSMUST00000233818.2 ENSMUST00000233945.2 ENSMUST00000166952.8 |
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein |
| chr6_-_52217821 | 6.26 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr6_-_52141796 | 6.04 |
ENSMUST00000014848.11
|
Hoxa2
|
homeobox A2 |
| chr16_+_96001650 | 5.70 |
ENSMUST00000048770.16
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein |
| chr13_-_63006176 | 5.69 |
ENSMUST00000021907.9
|
Fbp2
|
fructose bisphosphatase 2 |
| chr17_+_35295909 | 5.68 |
ENSMUST00000013910.5
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr14_-_61283911 | 5.57 |
ENSMUST00000111234.10
ENSMUST00000224371.2 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr11_+_96820220 | 5.38 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr14_+_102077937 | 5.26 |
ENSMUST00000159026.8
|
Lmo7
|
LIM domain only 7 |
| chr17_+_35295894 | 5.07 |
ENSMUST00000172678.8
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr3_-_30563831 | 5.02 |
ENSMUST00000173495.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr6_-_52222776 | 4.91 |
ENSMUST00000048026.10
|
Hoxa11
|
homeobox A11 |
| chr3_-_30563919 | 4.75 |
ENSMUST00000172697.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr10_+_43455157 | 4.73 |
ENSMUST00000058714.10
|
Cd24a
|
CD24a antigen |
| chr8_+_89247976 | 4.69 |
ENSMUST00000034086.13
|
Nkd1
|
naked cuticle 1 |
| chr11_+_96820091 | 4.54 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr3_-_141637245 | 4.47 |
ENSMUST00000106232.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chrX_+_162923474 | 4.40 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr17_+_48037758 | 4.36 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr6_-_52211882 | 4.29 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr14_+_101967343 | 4.22 |
ENSMUST00000100337.10
|
Lmo7
|
LIM domain only 7 |
| chr3_-_30563611 | 4.21 |
ENSMUST00000173899.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr5_+_17779721 | 4.16 |
ENSMUST00000169603.2
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr5_+_31409021 | 4.06 |
ENSMUST00000054829.13
ENSMUST00000201625.4 ENSMUST00000201937.4 |
Krtcap3
|
keratinocyte associated protein 3 |
| chr10_+_79658392 | 3.96 |
ENSMUST00000219305.2
ENSMUST00000046833.5 ENSMUST00000218687.2 |
Misp
|
mitotic spindle positioning |
| chr4_-_132990362 | 3.90 |
ENSMUST00000105908.10
ENSMUST00000030674.8 |
Sytl1
|
synaptotagmin-like 1 |
| chr8_-_11362731 | 3.87 |
ENSMUST00000033898.10
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr8_+_11362805 | 3.79 |
ENSMUST00000033899.14
|
Col4a2
|
collagen, type IV, alpha 2 |
| chr2_+_138120401 | 3.73 |
ENSMUST00000075410.5
|
Btbd3
|
BTB (POZ) domain containing 3 |
| chr2_+_70305267 | 3.70 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5 |
| chr14_+_56062422 | 3.68 |
ENSMUST00000172271.9
|
Nfatc4
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
| chr19_+_55730696 | 3.64 |
ENSMUST00000153888.9
ENSMUST00000127233.9 ENSMUST00000061496.17 ENSMUST00000111656.8 ENSMUST00000111657.11 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr2_-_115895528 | 3.58 |
ENSMUST00000028639.13
ENSMUST00000102538.11 |
Meis2
|
Meis homeobox 2 |
| chr3_-_89245297 | 3.57 |
ENSMUST00000029674.8
|
Efna4
|
ephrin A4 |
| chr16_+_96001915 | 3.56 |
ENSMUST00000132424.9
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein |
| chr15_+_10952418 | 3.53 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr11_+_96214078 | 3.52 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr14_+_56062252 | 3.51 |
ENSMUST00000024179.6
|
Nfatc4
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
| chr13_-_113182891 | 3.51 |
ENSMUST00000231962.2
ENSMUST00000022282.6 |
Gpx8
|
glutathione peroxidase 8 (putative) |
| chr6_+_15185399 | 3.46 |
ENSMUST00000115474.8
ENSMUST00000115472.8 ENSMUST00000031545.14 |
Foxp2
|
forkhead box P2 |
| chr15_-_100585789 | 3.38 |
ENSMUST00000023775.9
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr6_+_21985902 | 3.36 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr6_+_86605146 | 3.28 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr12_+_76118790 | 3.25 |
ENSMUST00000131480.8
|
Syne2
|
spectrin repeat containing, nuclear envelope 2 |
| chrX_+_72719098 | 3.24 |
ENSMUST00000171398.2
|
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr6_-_148846247 | 3.23 |
ENSMUST00000111562.8
ENSMUST00000081956.12 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr1_-_79417732 | 3.22 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr17_+_35296056 | 3.20 |
ENSMUST00000172959.2
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr17_+_35295849 | 3.20 |
ENSMUST00000172494.8
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr6_+_17065141 | 3.16 |
ENSMUST00000115467.11
ENSMUST00000154266.3 ENSMUST00000076654.9 |
Tes
|
testin LIM domain protein |
| chr11_+_108811626 | 3.15 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr2_-_173118315 | 3.14 |
ENSMUST00000036248.13
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_+_29735666 | 3.08 |
ENSMUST00000001812.5
|
Smo
|
smoothened, frizzled class receptor |
| chr1_+_131671751 | 3.07 |
ENSMUST00000049027.10
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr11_+_96183294 | 3.05 |
ENSMUST00000173432.3
|
Hoxb6
|
homeobox B6 |
| chr11_+_73068063 | 3.04 |
ENSMUST00000108477.2
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr2_-_163239865 | 3.04 |
ENSMUST00000017961.11
ENSMUST00000109425.3 |
Jph2
|
junctophilin 2 |
| chr4_-_141325517 | 3.03 |
ENSMUST00000131317.8
ENSMUST00000006381.11 ENSMUST00000129602.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr2_+_74552322 | 3.01 |
ENSMUST00000047904.4
|
Hoxd4
|
homeobox D4 |
| chr12_+_29988035 | 3.01 |
ENSMUST00000122328.8
ENSMUST00000118321.3 |
Pxdn
|
peroxidasin |
| chr2_+_103254465 | 2.98 |
ENSMUST00000171693.8
|
Elf5
|
E74-like factor 5 |
| chr10_+_38841511 | 2.97 |
ENSMUST00000019992.6
|
Lama4
|
laminin, alpha 4 |
| chr11_+_73067909 | 2.95 |
ENSMUST00000040687.12
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr19_-_38031774 | 2.94 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr16_+_78727829 | 2.92 |
ENSMUST00000114216.2
ENSMUST00000069148.13 ENSMUST00000023568.14 |
Chodl
|
chondrolectin |
| chr12_-_57592907 | 2.86 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr5_-_147244074 | 2.83 |
ENSMUST00000031650.4
|
Cdx2
|
caudal type homeobox 2 |
| chr2_+_103254401 | 2.83 |
ENSMUST00000028609.14
|
Elf5
|
E74-like factor 5 |
| chr11_+_108811168 | 2.82 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr10_+_21758083 | 2.80 |
ENSMUST00000120509.8
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_+_85972125 | 2.80 |
ENSMUST00000107178.9
ENSMUST00000048885.12 ENSMUST00000141889.8 ENSMUST00000120678.2 |
Adamtsl1
|
ADAMTS-like 1 |
| chr7_+_100143250 | 2.78 |
ENSMUST00000153287.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr3_-_141687987 | 2.76 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr18_-_61169262 | 2.74 |
ENSMUST00000025521.9
|
Cdx1
|
caudal type homeobox 1 |
| chr18_+_35347983 | 2.74 |
ENSMUST00000235449.2
ENSMUST00000235269.2 |
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr16_-_4950285 | 2.70 |
ENSMUST00000035672.5
|
Ppl
|
periplakin |
| chr3_+_89122499 | 2.64 |
ENSMUST00000142051.8
ENSMUST00000119084.2 |
Thbs3
|
thrombospondin 3 |
| chr1_-_186437760 | 2.61 |
ENSMUST00000195201.2
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr3_+_89122477 | 2.59 |
ENSMUST00000029682.11
|
Thbs3
|
thrombospondin 3 |
| chr17_-_71158184 | 2.54 |
ENSMUST00000059775.15
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr18_+_21205386 | 2.48 |
ENSMUST00000082235.5
|
Mep1b
|
meprin 1 beta |
| chr4_+_47208004 | 2.38 |
ENSMUST00000082303.13
ENSMUST00000102917.11 |
Col15a1
|
collagen, type XV, alpha 1 |
| chr11_-_86964881 | 2.38 |
ENSMUST00000020804.8
|
Gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr16_+_95058417 | 2.37 |
ENSMUST00000113861.8
ENSMUST00000113854.8 ENSMUST00000113862.8 ENSMUST00000037154.14 ENSMUST00000113855.8 |
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr17_-_68311073 | 2.37 |
ENSMUST00000024840.12
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr8_+_45960804 | 2.35 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_+_74557418 | 2.35 |
ENSMUST00000111980.4
|
Hoxd4
|
homeobox D4 |
| chrX_+_157993303 | 2.33 |
ENSMUST00000112493.8
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr4_+_41465134 | 2.32 |
ENSMUST00000030154.7
|
Nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr13_-_72111832 | 2.31 |
ENSMUST00000077337.9
|
Irx1
|
Iroquois homeobox 1 |
| chr2_-_34261121 | 2.30 |
ENSMUST00000127353.3
ENSMUST00000141653.3 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr12_+_76593799 | 2.29 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr2_-_173117936 | 2.29 |
ENSMUST00000139306.2
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_+_21986445 | 2.28 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr18_+_11972277 | 2.27 |
ENSMUST00000171109.9
ENSMUST00000046948.10 |
Cables1
|
CDK5 and Abl enzyme substrate 1 |
| chr13_+_118851214 | 2.26 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr15_-_50753061 | 2.26 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr10_-_85847697 | 2.25 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr16_+_44913974 | 2.24 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr3_-_51468236 | 2.23 |
ENSMUST00000037141.9
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr3_+_8574420 | 2.23 |
ENSMUST00000029002.9
|
Stmn2
|
stathmin-like 2 |
| chr1_-_186438177 | 2.22 |
ENSMUST00000045288.14
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr17_-_48716756 | 2.19 |
ENSMUST00000160319.8
ENSMUST00000159535.2 ENSMUST00000078800.13 ENSMUST00000046719.14 ENSMUST00000162460.8 |
Nfya
|
nuclear transcription factor-Y alpha |
| chr2_+_31840340 | 2.17 |
ENSMUST00000148056.4
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr6_-_94260806 | 2.16 |
ENSMUST00000203519.3
ENSMUST00000204347.3 |
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr10_-_12745109 | 2.16 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr19_+_55730316 | 2.14 |
ENSMUST00000111658.10
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr17_-_71158052 | 2.13 |
ENSMUST00000186358.6
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr4_-_94538370 | 2.13 |
ENSMUST00000053419.9
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr4_+_117706390 | 2.12 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_+_100142977 | 2.12 |
ENSMUST00000129324.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr18_-_15536747 | 2.11 |
ENSMUST00000079081.8
|
Aqp4
|
aquaporin 4 |
| chr16_+_44914397 | 2.10 |
ENSMUST00000061050.6
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr16_+_95058505 | 2.10 |
ENSMUST00000113856.8
ENSMUST00000125847.2 ENSMUST00000134166.8 ENSMUST00000140222.8 |
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_+_43672856 | 2.09 |
ENSMUST00000076939.10
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr19_+_55730488 | 2.06 |
ENSMUST00000111659.9
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr14_-_55344004 | 2.05 |
ENSMUST00000036041.15
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit |
| chr4_-_136563154 | 2.02 |
ENSMUST00000105846.9
ENSMUST00000059287.14 ENSMUST00000105845.9 |
Ephb2
|
Eph receptor B2 |
| chr14_-_98406977 | 2.00 |
ENSMUST00000071533.13
ENSMUST00000069334.8 |
Dach1
|
dachshund family transcription factor 1 |
| chr11_-_76462353 | 2.00 |
ENSMUST00000072740.13
|
Abr
|
active BCR-related gene |
| chr3_-_57483175 | 1.98 |
ENSMUST00000029380.14
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr14_+_70791496 | 1.97 |
ENSMUST00000022691.14
|
Hr
|
lysine demethylase and nuclear receptor corepressor |
| chr6_-_52195663 | 1.96 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr9_+_72600721 | 1.96 |
ENSMUST00000238315.2
|
Nedd4
|
neural precursor cell expressed, developmentally down-regulated 4 |
| chr2_-_65397809 | 1.94 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr2_-_65397850 | 1.93 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr4_+_62883796 | 1.91 |
ENSMUST00000030043.13
ENSMUST00000107415.8 ENSMUST00000064814.6 |
Zfp618
|
zinc finger protein 618 |
| chr13_+_5911481 | 1.90 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6 |
| chr1_+_107288928 | 1.90 |
ENSMUST00000191425.7
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr8_+_45960931 | 1.89 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_-_77380492 | 1.89 |
ENSMUST00000037593.14
ENSMUST00000092892.10 |
Ankrd13b
|
ankyrin repeat domain 13b |
| chr3_-_75864195 | 1.88 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr5_+_57875309 | 1.86 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr2_+_31840151 | 1.86 |
ENSMUST00000001920.13
ENSMUST00000151276.3 |
Aif1l
|
allograft inflammatory factor 1-like |
| chr10_+_69761597 | 1.86 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr2_+_74535242 | 1.85 |
ENSMUST00000019749.4
|
Hoxd8
|
homeobox D8 |
| chr3_-_57483330 | 1.84 |
ENSMUST00000120977.2
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr7_-_144493560 | 1.84 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr14_-_50020788 | 1.83 |
ENSMUST00000118129.2
ENSMUST00000036972.14 |
Armh4
|
armadillo-like helical domain containing 4 |
| chr4_+_117706559 | 1.83 |
ENSMUST00000163288.2
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_+_157401998 | 1.80 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr12_+_21161779 | 1.79 |
ENSMUST00000050990.10
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr3_-_27950491 | 1.79 |
ENSMUST00000058077.4
|
Tmem212
|
transmembrane protein 212 |
| chr6_+_48963795 | 1.78 |
ENSMUST00000037696.6
|
Svs1
|
seminal vesicle secretory protein 1 |
| chr15_-_54141816 | 1.78 |
ENSMUST00000079772.4
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr3_-_24837772 | 1.77 |
ENSMUST00000203414.2
|
Naaladl2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr2_+_174602412 | 1.74 |
ENSMUST00000029030.9
|
Edn3
|
endothelin 3 |
| chr5_+_53748323 | 1.73 |
ENSMUST00000201883.4
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_-_47270201 | 1.73 |
ENSMUST00000077221.6
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr11_+_87959067 | 1.71 |
ENSMUST00000018521.11
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr12_-_16850674 | 1.71 |
ENSMUST00000162112.8
|
Greb1
|
gene regulated by estrogen in breast cancer protein |
| chr1_-_181847492 | 1.70 |
ENSMUST00000177811.8
ENSMUST00000111025.8 ENSMUST00000111024.10 |
Enah
|
ENAH actin regulator |
| chr6_+_125016723 | 1.69 |
ENSMUST00000140131.8
ENSMUST00000032480.14 |
Ing4
|
inhibitor of growth family, member 4 |
| chr19_-_28657477 | 1.69 |
ENSMUST00000162022.8
ENSMUST00000112612.9 |
Glis3
|
GLIS family zinc finger 3 |
| chr2_-_57004933 | 1.69 |
ENSMUST00000028166.9
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr13_-_101831020 | 1.69 |
ENSMUST00000185795.2
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr19_+_44980565 | 1.68 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr2_+_74505350 | 1.67 |
ENSMUST00000001878.6
|
Hoxd12
|
homeobox D12 |
| chr4_-_58499398 | 1.67 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr10_+_69761784 | 1.66 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr8_+_93628015 | 1.65 |
ENSMUST00000104947.5
|
Capns2
|
calpain, small subunit 2 |
| chr5_-_148329615 | 1.65 |
ENSMUST00000138257.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr19_+_55730242 | 1.65 |
ENSMUST00000111662.11
ENSMUST00000041717.14 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr3_+_7568481 | 1.65 |
ENSMUST00000051064.9
ENSMUST00000193010.2 |
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr13_-_105191403 | 1.65 |
ENSMUST00000063551.7
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein |
| chr8_+_79754980 | 1.63 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr10_+_102348076 | 1.62 |
ENSMUST00000219445.2
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr10_-_37014859 | 1.62 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr3_+_54063459 | 1.60 |
ENSMUST00000029311.11
ENSMUST00000200048.5 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr11_-_101676076 | 1.57 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr16_+_95058895 | 1.57 |
ENSMUST00000113859.8
ENSMUST00000152516.2 |
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr7_-_83533497 | 1.57 |
ENSMUST00000094216.5
|
Tlnrd1
|
talin rod domain containing 1 |
| chr6_+_83011154 | 1.56 |
ENSMUST00000000707.9
ENSMUST00000101257.4 |
Loxl3
|
lysyl oxidase-like 3 |
| chr15_+_102922247 | 1.56 |
ENSMUST00000001709.3
|
Hoxc5
|
homeobox C5 |
| chr11_-_106606076 | 1.56 |
ENSMUST00000080853.11
ENSMUST00000183610.8 ENSMUST00000103069.10 ENSMUST00000106796.9 |
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr9_-_71803354 | 1.56 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr11_+_108814007 | 1.55 |
ENSMUST00000106711.2
|
Axin2
|
axin 2 |
| chrX_+_108138965 | 1.53 |
ENSMUST00000033598.9
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr15_+_102898966 | 1.52 |
ENSMUST00000001703.8
|
Hoxc8
|
homeobox C8 |
| chr1_+_187730032 | 1.51 |
ENSMUST00000110938.2
|
Esrrg
|
estrogen-related receptor gamma |
| chr12_+_80565764 | 1.46 |
ENSMUST00000021558.8
|
Galnt16
|
polypeptide N-acetylgalactosaminyltransferase 16 |
| chr2_-_75534985 | 1.46 |
ENSMUST00000102672.5
|
Nfe2l2
|
nuclear factor, erythroid derived 2, like 2 |
| chr19_-_56996617 | 1.46 |
ENSMUST00000118800.8
ENSMUST00000111584.9 ENSMUST00000122359.8 ENSMUST00000148049.8 |
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr11_-_106605772 | 1.45 |
ENSMUST00000124958.3
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr9_-_44646487 | 1.45 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr6_-_72766224 | 1.44 |
ENSMUST00000069536.12
|
Tcf7l1
|
transcription factor 7 like 1 (T cell specific, HMG box) |
| chr6_-_72765935 | 1.44 |
ENSMUST00000114053.9
|
Tcf7l1
|
transcription factor 7 like 1 (T cell specific, HMG box) |
| chr18_+_4921663 | 1.42 |
ENSMUST00000143254.8
|
Svil
|
supervillin |
| chr17_+_48717007 | 1.41 |
ENSMUST00000167180.8
ENSMUST00000046651.7 ENSMUST00000233426.2 |
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr9_-_117701613 | 1.39 |
ENSMUST00000239475.2
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr2_-_170248421 | 1.39 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr13_-_101829070 | 1.39 |
ENSMUST00000187009.7
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.6 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 2.7 | 8.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 1.8 | 7.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.6 | 4.8 | GO:1905006 | positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 1.6 | 4.7 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.5 | 6.0 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 1.4 | 7.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.4 | 8.6 | GO:2000054 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 1.3 | 3.9 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.3 | 5.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 1.1 | 11.3 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 1.1 | 4.4 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 1.1 | 3.2 | GO:0015881 | creatine transport(GO:0015881) |
| 1.0 | 3.0 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 1.0 | 2.9 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 1.0 | 2.9 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.9 | 5.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.9 | 14.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.9 | 4.4 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.8 | 5.8 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.8 | 6.9 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.8 | 3.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.8 | 2.3 | GO:0060447 | bronchiole development(GO:0060435) bud outgrowth involved in lung branching(GO:0060447) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.7 | 3.6 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.7 | 2.0 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.7 | 6.6 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.7 | 1.3 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.6 | 3.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.6 | 1.3 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.6 | 2.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.6 | 3.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.6 | 4.7 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.6 | 2.3 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.6 | 2.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.5 | 1.6 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.5 | 4.2 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.5 | 1.5 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.5 | 3.4 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.5 | 6.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.5 | 2.7 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.5 | 2.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.4 | 3.5 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.4 | 15.4 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.4 | 5.2 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.4 | 1.3 | GO:1903210 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.4 | 15.9 | GO:0060065 | uterus development(GO:0060065) |
| 0.4 | 3.9 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.4 | 5.6 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.4 | 1.3 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.4 | 2.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.4 | 1.7 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.4 | 2.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.4 | 1.7 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.4 | 1.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.4 | 3.3 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.4 | 2.4 | GO:0035128 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.4 | 1.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.4 | 3.8 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.4 | 3.0 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.4 | 2.2 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.3 | 1.7 | GO:0060853 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) regulation of cell adhesion involved in heart morphogenesis(GO:0061344) blood vessel endothelial cell fate specification(GO:0097101) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.3 | 2.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.3 | 1.3 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.3 | 1.3 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.3 | 2.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.3 | 4.3 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.3 | 0.9 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.3 | 7.5 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.3 | 2.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 2.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 0.8 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.3 | 1.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 0.8 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.3 | 1.8 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 | 0.7 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.2 | 1.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 1.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.2 | 1.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 | 0.7 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.2 | 0.9 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.2 | 0.8 | GO:0009196 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.2 | 4.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 1.0 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 0.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 3.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 1.6 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 | 2.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 3.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 2.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.6 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.2 | 1.8 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.2 | 4.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 0.5 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.2 | 2.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 4.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 2.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 0.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 0.8 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 1.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.2 | 2.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 2.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 4.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 0.8 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 | 3.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 2.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.1 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 3.5 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 1.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.4 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.1 | 1.8 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 | 0.6 | GO:0046016 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) positive regulation of transcription by glucose(GO:0046016) |
| 0.1 | 1.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 5.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 1.8 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.9 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 2.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 2.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 3.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.9 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 6.0 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.7 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 2.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 1.7 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 3.0 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 1.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 4.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 2.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 1.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.9 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 3.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 14.0 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 1.6 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 0.3 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 0.7 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.1 | 0.8 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.6 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 2.4 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.1 | 1.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.2 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.5 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.3 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.8 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 0.7 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 1.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0061299 | extracellular matrix-cell signaling(GO:0035426) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 2.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 3.4 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 3.4 | GO:0001942 | hair follicle development(GO:0001942) skin epidermis development(GO:0098773) |
| 0.0 | 1.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.5 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.5 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.9 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 1.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 1.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.0 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.7 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 1.7 | GO:2000648 | positive regulation of stem cell proliferation(GO:2000648) |
| 0.0 | 1.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 4.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.5 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.7 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.4 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.0 | 0.3 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 1.8 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 2.5 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 1.6 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.5 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 1.3 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.6 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.2 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.5 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.2 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 1.4 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 2.4 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0090102 | cochlea development(GO:0090102) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.9 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 1.1 | 11.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 1.1 | 7.7 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 1.1 | 4.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.4 | 1.3 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.4 | 2.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.4 | 2.4 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.3 | 4.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 3.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 3.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 1.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 1.6 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.3 | 2.9 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 1.0 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.2 | 1.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.2 | 6.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 1.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 6.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 3.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 1.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 4.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 2.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 0.8 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.9 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 4.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 4.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 4.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 1.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 18.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 13.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 2.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.3 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 3.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 9.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.9 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 5.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 7.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 7.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.5 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 4.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 30.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 9.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 8.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 3.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 8.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 13.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.6 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 5.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 2.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 2.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 29.6 | GO:0070062 | extracellular exosome(GO:0070062) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 17.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.1 | 3.2 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 1.0 | 7.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.8 | 4.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.8 | 4.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.7 | 4.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.7 | 4.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.6 | 7.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.6 | 2.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 9.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.5 | 16.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 1.7 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.4 | 1.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.4 | 1.9 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.4 | 2.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.3 | 2.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 1.0 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.3 | 2.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 7.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 2.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 2.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 1.0 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 0.9 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.3 | 2.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 5.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.3 | 3.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.3 | 8.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 6.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 0.8 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.3 | 1.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 2.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 3.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 3.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 7.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 2.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 4.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.9 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.2 | 0.8 | GO:0004127 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.2 | 1.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.2 | 2.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 5.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 1.7 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 1.3 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.2 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 4.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 4.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 3.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 3.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 7.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.2 | 1.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 2.9 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 14.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 2.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 0.3 | GO:0015116 | sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 4.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 4.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 3.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 6.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 3.9 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.9 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 4.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 2.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 2.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.6 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 2.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 2.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 16.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 1.7 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 6.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 4.3 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 0.4 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.0 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 2.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 10.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 2.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 4.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 3.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 5.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 5.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 7.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) triplex DNA binding(GO:0045142) |
| 0.0 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 7.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.8 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 15.0 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 11.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 8.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 6.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 14.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 14.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 9.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 6.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 8.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 12.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 7.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 15.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 6.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 6.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 11.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 9.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 10.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 3.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 6.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 2.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 6.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 5.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 3.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 6.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 17.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 2.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 4.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 5.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 6.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 4.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 3.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 5.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 4.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 4.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 3.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 2.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 3.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.5 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |