Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
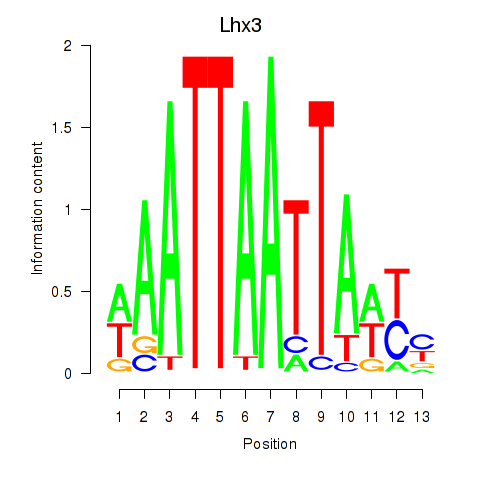
Results for Lhx3
Z-value: 0.47
Transcription factors associated with Lhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx3
|
ENSMUSG00000026934.16 | Lhx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx3 | mm39_v1_chr2_-_26096547_26096592 | -0.06 | 6.0e-01 | Click! |
Activity profile of Lhx3 motif
Sorted Z-values of Lhx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_39801188 | 3.35 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr3_+_59989282 | 3.21 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr19_-_39875192 | 2.71 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr3_+_132335575 | 2.24 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr7_-_44753168 | 2.19 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr3_+_132335704 | 2.15 |
ENSMUST00000212594.2
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr2_+_110427643 | 2.13 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr17_-_36343573 | 1.92 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr6_-_115569504 | 1.85 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr9_-_110576124 | 1.75 |
ENSMUST00000199862.5
ENSMUST00000198865.5 |
Pth1r
|
parathyroid hormone 1 receptor |
| chr11_+_76792977 | 1.73 |
ENSMUST00000102495.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr17_-_35081456 | 1.61 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr17_-_35081129 | 1.60 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr3_+_122213420 | 1.60 |
ENSMUST00000029766.9
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr5_-_51711237 | 1.57 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chrM_+_9870 | 1.51 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr5_-_51711204 | 1.49 |
ENSMUST00000196968.5
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr6_-_70116066 | 1.39 |
ENSMUST00000103379.3
ENSMUST00000197371.2 |
Igkv6-29
|
immunoglobulin kappa chain variable 6-29 |
| chr1_-_36312482 | 1.36 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr11_+_109434519 | 1.35 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr6_-_70313491 | 1.33 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr18_-_38999755 | 1.31 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr9_-_110576192 | 1.20 |
ENSMUST00000199791.2
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr3_-_14843512 | 1.19 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr1_+_21310803 | 1.19 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr1_+_21310821 | 1.17 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chrM_+_9459 | 1.10 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr9_-_15212745 | 1.08 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr3_+_95226093 | 1.08 |
ENSMUST00000139866.2
|
Cers2
|
ceramide synthase 2 |
| chr10_+_101994841 | 1.07 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr11_+_58311921 | 1.07 |
ENSMUST00000013797.3
|
1810065E05Rik
|
RIKEN cDNA 1810065E05 gene |
| chr10_+_101994719 | 0.99 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr7_-_45480200 | 0.97 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr11_-_106205320 | 0.91 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr18_-_39000056 | 0.77 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr14_+_80237691 | 0.77 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr2_+_22959223 | 0.76 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr13_-_101829070 | 0.76 |
ENSMUST00000187009.7
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr9_+_21634779 | 0.73 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr16_+_11224481 | 0.73 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chrM_+_14138 | 0.72 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr6_-_3399451 | 0.69 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr3_+_151143524 | 0.69 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr10_+_5543769 | 0.68 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr9_+_120758282 | 0.65 |
ENSMUST00000130466.8
|
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr14_-_70945434 | 0.65 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr12_-_25147139 | 0.65 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr8_+_22329942 | 0.64 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2 |
| chr7_-_119058489 | 0.63 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr16_+_45044678 | 0.63 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr11_+_70410445 | 0.63 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr15_+_65682066 | 0.63 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr2_+_22959452 | 0.60 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr17_-_78991691 | 0.59 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr3_-_144514386 | 0.59 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr12_-_84664001 | 0.55 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr10_-_8638215 | 0.55 |
ENSMUST00000212553.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr10_-_62343516 | 0.50 |
ENSMUST00000020271.13
|
Srgn
|
serglycin |
| chr15_-_101422054 | 0.49 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chrM_-_14061 | 0.48 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr3_+_151143557 | 0.48 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr13_-_101829132 | 0.48 |
ENSMUST00000035532.13
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr3_-_67422821 | 0.48 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr4_-_3938352 | 0.48 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr1_+_177272297 | 0.48 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr8_+_84262409 | 0.46 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr14_+_53994813 | 0.44 |
ENSMUST00000180380.3
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr5_+_117378510 | 0.43 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr11_-_4045343 | 0.40 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr6_-_50631418 | 0.40 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr14_+_53310220 | 0.39 |
ENSMUST00000196079.2
|
Trav13d-4
|
T cell receptor alpha variable 13D-4 |
| chr13_+_93440572 | 0.39 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr14_-_48902555 | 0.38 |
ENSMUST00000118578.9
|
Otx2
|
orthodenticle homeobox 2 |
| chr7_+_99808452 | 0.38 |
ENSMUST00000032967.4
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr17_+_79919267 | 0.38 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chrX_-_138772383 | 0.37 |
ENSMUST00000033811.14
ENSMUST00000087401.12 |
Morc4
|
microrchidia 4 |
| chr7_-_115459082 | 0.37 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr4_+_100336003 | 0.37 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr13_+_19526322 | 0.37 |
ENSMUST00000184430.2
|
Trgj4
|
T cell receptor gamma joining 4 |
| chr2_+_36120438 | 0.36 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr1_-_166509404 | 0.36 |
ENSMUST00000148677.2
ENSMUST00000027843.11 |
Fmo9
|
flavin containing monooxygenase 9 |
| chr10_+_36382810 | 0.36 |
ENSMUST00000167191.8
ENSMUST00000058738.11 |
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chrX_-_142716085 | 0.36 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr18_+_20569226 | 0.33 |
ENSMUST00000019426.5
|
Dsg4
|
desmoglein 4 |
| chr7_+_126575752 | 0.33 |
ENSMUST00000206346.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr5_-_62923463 | 0.32 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chrX_-_8118541 | 0.32 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr11_-_101066266 | 0.31 |
ENSMUST00000062759.4
|
Ccr10
|
chemokine (C-C motif) receptor 10 |
| chr11_-_99265721 | 0.30 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr1_-_37955569 | 0.29 |
ENSMUST00000078307.7
|
Lyg2
|
lysozyme G-like 2 |
| chr1_+_177272215 | 0.28 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr2_+_3115250 | 0.27 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr17_+_38104420 | 0.26 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr1_+_153541339 | 0.26 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr2_+_81883566 | 0.24 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr1_+_153541412 | 0.23 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr5_+_96357337 | 0.22 |
ENSMUST00000117766.8
|
Mrpl1
|
mitochondrial ribosomal protein L1 |
| chr9_+_96140750 | 0.22 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr10_+_115979787 | 0.22 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chrX_+_41241049 | 0.22 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr10_+_23672842 | 0.21 |
ENSMUST00000119597.8
ENSMUST00000179321.8 ENSMUST00000133289.2 |
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr11_-_99979052 | 0.21 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr9_-_103569984 | 0.21 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chr15_-_103473481 | 0.21 |
ENSMUST00000228060.2
ENSMUST00000228895.2 ENSMUST00000023134.5 |
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr6_+_43083303 | 0.19 |
ENSMUST00000213649.2
|
Olfr441
|
olfactory receptor 441 |
| chr2_+_152468850 | 0.19 |
ENSMUST00000000369.4
ENSMUST00000150913.2 |
Rem1
|
rad and gem related GTP binding protein 1 |
| chrX_-_99669507 | 0.19 |
ENSMUST00000059099.7
|
Pdzd11
|
PDZ domain containing 11 |
| chr14_-_50558325 | 0.18 |
ENSMUST00000050928.3
|
Olfr734
|
olfactory receptor 734 |
| chr13_+_23343482 | 0.18 |
ENSMUST00000226845.2
ENSMUST00000228666.2 ENSMUST00000227388.2 |
Vmn1r219
|
vomeronasal 1 receptor 219 |
| chr10_+_74872898 | 0.18 |
ENSMUST00000147802.9
ENSMUST00000020391.13 ENSMUST00000234625.2 |
Rab36
|
RAB36, member RAS oncogene family |
| chr5_+_96357974 | 0.17 |
ENSMUST00000036437.13
ENSMUST00000121477.2 |
Mrpl1
|
mitochondrial ribosomal protein L1 |
| chr17_+_37977879 | 0.17 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chr11_+_73489420 | 0.17 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr9_+_96140781 | 0.16 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr2_+_163500290 | 0.16 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr11_-_49004584 | 0.15 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr7_+_43077088 | 0.15 |
ENSMUST00000239023.2
|
Gm38999
|
predicted gene, 38999 |
| chr11_-_30148230 | 0.14 |
ENSMUST00000102838.10
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr8_-_22396428 | 0.13 |
ENSMUST00000051965.5
|
Defb11
|
defensin beta 11 |
| chr8_-_3674993 | 0.13 |
ENSMUST00000142431.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr2_-_72817060 | 0.12 |
ENSMUST00000112062.2
|
Gm11084
|
predicted gene 11084 |
| chr17_-_37615204 | 0.12 |
ENSMUST00000214376.2
|
Olfr101
|
olfactory receptor 101 |
| chr17_-_14914484 | 0.12 |
ENSMUST00000170872.3
|
Thbs2
|
thrombospondin 2 |
| chr14_-_96756503 | 0.12 |
ENSMUST00000022666.9
|
Klhl1
|
kelch-like 1 |
| chr2_-_87570322 | 0.12 |
ENSMUST00000214573.2
|
Olfr1138
|
olfactory receptor 1138 |
| chr2_-_86257093 | 0.12 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr5_-_104225458 | 0.11 |
ENSMUST00000198485.5
ENSMUST00000164471.8 ENSMUST00000178967.2 |
Gm17660
|
predicted gene, 17660 |
| chr4_+_102446883 | 0.10 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr7_-_12829100 | 0.09 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr5_-_74863514 | 0.09 |
ENSMUST00000117388.8
|
Lnx1
|
ligand of numb-protein X 1 |
| chr7_+_20606181 | 0.09 |
ENSMUST00000164288.2
|
Vmn1r116
|
vomeronasal 1 receptor 116 |
| chr10_+_129601351 | 0.09 |
ENSMUST00000203236.3
|
Olfr808
|
olfactory receptor 808 |
| chr18_-_42132106 | 0.09 |
ENSMUST00000097591.5
|
Grxcr2
|
glutaredoxin, cysteine rich 2 |
| chr3_+_41697046 | 0.08 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr17_+_37769807 | 0.07 |
ENSMUST00000214668.2
ENSMUST00000217602.2 ENSMUST00000214938.2 |
Olfr109
|
olfactory receptor 109 |
| chr2_-_89779008 | 0.07 |
ENSMUST00000214846.2
|
Olfr1259
|
olfactory receptor 1259 |
| chr5_-_3697806 | 0.07 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr7_-_103191732 | 0.06 |
ENSMUST00000215663.2
|
Olfr612
|
olfactory receptor 612 |
| chr6_+_37847721 | 0.06 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr7_+_102526329 | 0.06 |
ENSMUST00000098216.2
|
Olfr568
|
olfactory receptor 568 |
| chrX_+_162923474 | 0.06 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr9_+_111014254 | 0.06 |
ENSMUST00000198986.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr10_-_33500583 | 0.06 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr2_-_76700830 | 0.05 |
ENSMUST00000138542.2
|
Ttn
|
titin |
| chr14_+_8283087 | 0.04 |
ENSMUST00000206298.3
ENSMUST00000216079.2 |
Olfr720
|
olfactory receptor 720 |
| chr19_-_12313274 | 0.04 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr16_-_19443851 | 0.04 |
ENSMUST00000079891.4
|
Olfr171
|
olfactory receptor 171 |
| chr10_-_130265572 | 0.04 |
ENSMUST00000171811.4
|
Vmn2r85
|
vomeronasal 2, receptor 85 |
| chrX_+_65696608 | 0.04 |
ENSMUST00000036043.5
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr11_+_58485940 | 0.04 |
ENSMUST00000214990.2
ENSMUST00000216965.2 |
Olfr324
|
olfactory receptor 324 |
| chr11_+_58668915 | 0.03 |
ENSMUST00000081533.5
|
Olfr315
|
olfactory receptor 315 |
| chr9_+_18320390 | 0.03 |
ENSMUST00000098973.3
|
Ubtfl1
|
upstream binding transcription factor, RNA polymerase I-like 1 |
| chr9_+_40092216 | 0.03 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr17_+_69746321 | 0.03 |
ENSMUST00000169935.2
|
Akain1
|
A kinase (PRKA) anchor inhibitor 1 |
| chr2_+_87726521 | 0.03 |
ENSMUST00000052300.2
|
Olfr1153
|
olfactory receptor 1153 |
| chr14_+_53574579 | 0.02 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr14_+_51648458 | 0.02 |
ENSMUST00000022438.12
|
Vmn2r88
|
vomeronasal 2, receptor 88 |
| chr3_-_64473251 | 0.02 |
ENSMUST00000176481.9
|
Vmn2r6
|
vomeronasal 2, receptor 6 |
| chr14_-_109151590 | 0.01 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr15_+_98350469 | 0.01 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr4_+_147390131 | 0.01 |
ENSMUST00000148762.4
|
Zfp988
|
zinc finger protein 988 |
| chr13_+_83723743 | 0.01 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr4_+_145241454 | 0.01 |
ENSMUST00000105741.2
|
Zfp990
|
zinc finger protein 990 |
| chr4_-_146993984 | 0.01 |
ENSMUST00000238583.2
ENSMUST00000049821.4 |
Gm21411
|
predicted gene, 21411 |
| chr13_+_93441307 | 0.01 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr7_-_5325456 | 0.01 |
ENSMUST00000207520.2
|
Nlrp2
|
NLR family, pyrin domain containing 2 |
| chr12_-_118930130 | 0.01 |
ENSMUST00000035515.5
|
Abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr4_-_147894245 | 0.00 |
ENSMUST00000105734.10
ENSMUST00000176201.2 |
Zfp984
Gm20707
|
zinc finger protein 984 predicted gene 20707 |
| chr15_-_98507913 | 0.00 |
ENSMUST00000226500.2
ENSMUST00000227501.2 |
Adcy6
|
adenylate cyclase 6 |
| chr7_+_102549549 | 0.00 |
ENSMUST00000098215.3
|
Olfr570
|
olfactory receptor 570 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1904633 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.8 | 2.4 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.6 | 1.9 | GO:2000566 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.3 | 2.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 | 1.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 0.6 | GO:1904499 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.2 | 0.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 0.6 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.2 | 3.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 6.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 0.7 | GO:1903978 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) regulation of microglial cell activation(GO:1903978) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 2.9 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.4 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.1 | 0.5 | GO:0033373 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.1 | 1.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.7 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 1.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.7 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.4 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.4 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.4 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 1.0 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.4 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.6 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 1.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.5 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.7 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.3 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.8 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 0.6 | GO:0043276 | anoikis(GO:0043276) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.3 | 1.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 0.7 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 1.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.7 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.4 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 2.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 2.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 3.2 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.9 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 2.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 2.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 2.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 6.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 1.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 1.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.3 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 3.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 3.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.7 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 2.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 3.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |