Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Lhx4
Z-value: 0.86
Transcription factors associated with Lhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx4
|
ENSMUSG00000026468.15 | Lhx4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx4 | mm39_v1_chr1_-_155617773_155617790 | 0.16 | 1.8e-01 | Click! |
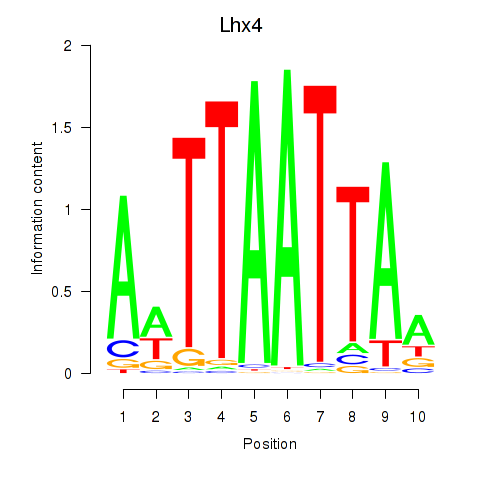
Activity profile of Lhx4 motif
Sorted Z-values of Lhx4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_44714263 | 8.61 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr11_-_46597885 | 7.78 |
ENSMUST00000055102.13
ENSMUST00000125008.2 |
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr7_-_14180496 | 6.54 |
ENSMUST00000063509.11
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chrX_+_149330371 | 6.54 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr7_+_30193047 | 5.45 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr17_+_12597490 | 4.66 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr19_-_39875192 | 4.26 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr5_-_87402659 | 4.07 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr19_-_39801188 | 4.05 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr12_-_25147139 | 3.94 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr19_+_30210320 | 3.73 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr4_+_95445731 | 3.63 |
ENSMUST00000079223.11
ENSMUST00000177394.8 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr17_-_35081456 | 3.40 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr1_-_72323464 | 3.34 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr17_-_35081129 | 3.30 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr19_+_37425180 | 3.24 |
ENSMUST00000128184.3
|
Hhex
|
hematopoietically expressed homeobox |
| chr1_-_72323407 | 3.21 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr2_-_134396268 | 3.16 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr9_+_108216466 | 3.07 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr6_-_141892517 | 3.04 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr9_-_110576192 | 2.93 |
ENSMUST00000199791.2
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr19_-_39637489 | 2.91 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr9_+_108216433 | 2.80 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr9_+_108216233 | 2.67 |
ENSMUST00000082429.8
|
Gpx1
|
glutathione peroxidase 1 |
| chr11_+_109434519 | 2.60 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr9_-_110576124 | 2.56 |
ENSMUST00000199862.5
ENSMUST00000198865.5 |
Pth1r
|
parathyroid hormone 1 receptor |
| chr9_+_65538352 | 2.48 |
ENSMUST00000216342.2
ENSMUST00000216382.2 |
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr8_+_22329942 | 2.41 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2 |
| chr7_-_12829100 | 2.32 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr4_+_100336003 | 2.27 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr1_-_162694076 | 2.22 |
ENSMUST00000046049.14
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr3_-_67422821 | 2.22 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr6_+_72074545 | 2.17 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr4_+_15881256 | 2.14 |
ENSMUST00000029876.2
|
Calb1
|
calbindin 1 |
| chr1_-_130589349 | 2.14 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr2_-_152422220 | 2.03 |
ENSMUST00000053180.4
|
Defb19
|
defensin beta 19 |
| chr4_-_19922599 | 2.02 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr14_+_26722319 | 1.92 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr5_+_130398261 | 1.87 |
ENSMUST00000086029.10
|
Caln1
|
calneuron 1 |
| chr2_+_22959223 | 1.85 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr11_+_32236945 | 1.82 |
ENSMUST00000101387.4
|
Hbq1b
|
hemoglobin, theta 1B |
| chr17_+_79919267 | 1.81 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr6_+_72074718 | 1.79 |
ENSMUST00000187007.3
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr4_-_45532470 | 1.76 |
ENSMUST00000147448.2
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr14_-_31139617 | 1.74 |
ENSMUST00000100730.10
|
Sh3bp5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr1_+_72323526 | 1.72 |
ENSMUST00000027380.12
ENSMUST00000141783.2 |
Tmem169
|
transmembrane protein 169 |
| chr7_+_30157704 | 1.70 |
ENSMUST00000126297.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr3_+_57332735 | 1.69 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr5_-_62923463 | 1.68 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_+_21437440 | 1.66 |
ENSMUST00000086361.12
ENSMUST00000173769.3 |
AB124611
|
cDNA sequence AB124611 |
| chr15_+_31224460 | 1.62 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr17_-_36343573 | 1.55 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr1_+_87983099 | 1.52 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr2_+_70305267 | 1.47 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5 |
| chr2_+_67935015 | 1.46 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr12_+_103524690 | 1.46 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr7_-_37472979 | 1.44 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr12_-_103660916 | 1.43 |
ENSMUST00000117053.8
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr2_-_63014622 | 1.41 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr1_+_88234454 | 1.41 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr7_+_45271229 | 1.39 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr15_+_31224616 | 1.39 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr9_-_15212745 | 1.38 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr4_+_114914880 | 1.33 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr4_+_34893772 | 1.31 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr1_+_153541339 | 1.31 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr8_-_22396428 | 1.29 |
ENSMUST00000051965.5
|
Defb11
|
defensin beta 11 |
| chr1_+_153541412 | 1.28 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr15_+_31225302 | 1.28 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr15_+_31224555 | 1.24 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr2_+_22959452 | 1.17 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr15_-_103356880 | 1.17 |
ENSMUST00000065978.9
|
Gtsf2
|
gametocyte specific factor 2 |
| chr1_+_132155922 | 1.14 |
ENSMUST00000191418.2
|
Lemd1
|
LEM domain containing 1 |
| chr1_+_87983189 | 1.13 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr3_+_68776884 | 1.11 |
ENSMUST00000054551.3
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chrX_-_142716085 | 1.10 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr3_+_122213420 | 1.09 |
ENSMUST00000029766.9
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr5_+_20112771 | 1.08 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_+_145397238 | 1.08 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr5_+_130413532 | 1.07 |
ENSMUST00000202728.2
|
Caln1
|
calneuron 1 |
| chr4_-_35845204 | 1.07 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr5_+_20112704 | 1.05 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr12_+_4004943 | 1.04 |
ENSMUST00000219543.2
ENSMUST00000218089.2 ENSMUST00000020990.7 ENSMUST00000218169.2 ENSMUST00000220006.2 |
Pomc
|
pro-opiomelanocortin-alpha |
| chr11_+_58062467 | 1.04 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr11_-_99979052 | 1.03 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr7_+_140521450 | 1.00 |
ENSMUST00000164580.3
ENSMUST00000079403.11 |
Pgghg
|
protein glucosylgalactosylhydroxylysine glucosidase |
| chr16_-_29363671 | 1.00 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr11_-_73217298 | 1.00 |
ENSMUST00000155630.9
|
Aspa
|
aspartoacylase |
| chr12_-_104439589 | 0.98 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr9_+_72714156 | 0.97 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chr2_+_81883566 | 0.96 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr2_+_124978518 | 0.96 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr2_-_164480710 | 0.96 |
ENSMUST00000109336.2
|
Wfdc16
|
WAP four-disulfide core domain 16 |
| chr18_-_39000056 | 0.96 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr1_+_19173246 | 0.94 |
ENSMUST00000037294.8
|
Tfap2d
|
transcription factor AP-2, delta |
| chr1_-_82564501 | 0.94 |
ENSMUST00000087050.7
|
Col4a4
|
collagen, type IV, alpha 4 |
| chr12_+_38830081 | 0.94 |
ENSMUST00000095767.11
|
Etv1
|
ets variant 1 |
| chr6_-_138404076 | 0.93 |
ENSMUST00000203435.3
|
Lmo3
|
LIM domain only 3 |
| chr12_+_38830283 | 0.91 |
ENSMUST00000162563.8
ENSMUST00000161164.8 ENSMUST00000160996.8 |
Etv1
|
ets variant 1 |
| chr16_+_43056218 | 0.91 |
ENSMUST00000146708.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr16_-_55103598 | 0.91 |
ENSMUST00000036412.4
|
Zpld1
|
zona pellucida like domain containing 1 |
| chr2_+_124978612 | 0.91 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr18_-_38999755 | 0.91 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr1_-_164988342 | 0.90 |
ENSMUST00000027859.12
|
Tbx19
|
T-box 19 |
| chr2_-_111779785 | 0.90 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr6_+_123239076 | 0.89 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr1_+_82564627 | 0.89 |
ENSMUST00000113457.9
|
Col4a3
|
collagen, type IV, alpha 3 |
| chr11_-_99494134 | 0.88 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr19_+_58500411 | 0.87 |
ENSMUST00000235305.2
ENSMUST00000069419.8 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr5_+_121601212 | 0.87 |
ENSMUST00000094357.11
ENSMUST00000031405.12 |
Tmem116
|
transmembrane protein 116 |
| chr6_+_86342622 | 0.86 |
ENSMUST00000071492.9
|
Fam136a
|
family with sequence similarity 136, member A |
| chr16_-_16647139 | 0.85 |
ENSMUST00000023468.6
|
Spag6l
|
sperm associated antigen 6-like |
| chr1_-_74924481 | 0.85 |
ENSMUST00000159232.2
ENSMUST00000068631.4 |
Fev
|
FEV transcription factor, ETS family member |
| chr2_-_32314017 | 0.84 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr4_-_82423985 | 0.83 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr12_+_38830812 | 0.83 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr2_+_61634797 | 0.82 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
| chrX_-_142716200 | 0.82 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr1_+_106099482 | 0.81 |
ENSMUST00000061047.7
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr11_+_46641330 | 0.81 |
ENSMUST00000109223.8
|
Havcr1
|
hepatitis A virus cellular receptor 1 |
| chr7_+_54485336 | 0.81 |
ENSMUST00000082373.8
|
Luzp2
|
leucine zipper protein 2 |
| chr17_+_45866618 | 0.80 |
ENSMUST00000024742.9
ENSMUST00000233929.2 |
Nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon |
| chr12_-_111947487 | 0.80 |
ENSMUST00000190536.2
|
Rd3l
|
retinal degeneration 3-like |
| chr7_-_49286594 | 0.78 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr14_+_27598021 | 0.77 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr14_+_65504067 | 0.77 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr7_-_44753168 | 0.76 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chrX_+_56013480 | 0.76 |
ENSMUST00000145035.2
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr6_-_50631418 | 0.76 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr1_-_9368721 | 0.76 |
ENSMUST00000132064.8
|
Sntg1
|
syntrophin, gamma 1 |
| chr2_-_63014514 | 0.73 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr16_-_22676264 | 0.72 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr18_-_42132106 | 0.72 |
ENSMUST00000097591.5
|
Grxcr2
|
glutaredoxin, cysteine rich 2 |
| chr17_+_29493049 | 0.71 |
ENSMUST00000149405.4
|
BC004004
|
cDNA sequence BC004004 |
| chr9_+_38755745 | 0.71 |
ENSMUST00000217350.2
|
Olfr924
|
olfactory receptor 924 |
| chr1_-_9369045 | 0.71 |
ENSMUST00000191683.6
|
Sntg1
|
syntrophin, gamma 1 |
| chr14_-_17760477 | 0.71 |
ENSMUST00000171305.2
ENSMUST00000169465.8 |
Gm7876
|
predicted gene 7876 |
| chr9_-_103569984 | 0.71 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chr8_+_66964401 | 0.71 |
ENSMUST00000002025.5
ENSMUST00000183187.2 |
Tktl2
|
transketolase-like 2 |
| chr14_+_53574579 | 0.70 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr9_+_118307412 | 0.70 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr19_-_20931566 | 0.69 |
ENSMUST00000039500.4
|
Tmc1
|
transmembrane channel-like gene family 1 |
| chr1_+_85503397 | 0.68 |
ENSMUST00000178024.2
|
G530012D18Rik
|
RIKEN cDNA G530012D1 gene |
| chr4_-_3938352 | 0.68 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr5_-_28415166 | 0.68 |
ENSMUST00000117098.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr3_-_86827640 | 0.67 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr6_+_34757346 | 0.66 |
ENSMUST00000115016.8
ENSMUST00000115017.8 |
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr5_+_20112500 | 0.66 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr17_+_70829050 | 0.65 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr4_+_146586445 | 0.65 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr19_+_44980565 | 0.64 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chrX_-_159689993 | 0.64 |
ENSMUST00000069417.6
|
Gja6
|
gap junction protein, alpha 6 |
| chr18_+_34891941 | 0.64 |
ENSMUST00000049281.12
|
Fam53c
|
family with sequence similarity 53, member C |
| chr5_-_28415361 | 0.64 |
ENSMUST00000141196.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr5_-_3697806 | 0.63 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr17_+_29493113 | 0.63 |
ENSMUST00000234326.2
ENSMUST00000235117.2 |
BC004004
|
cDNA sequence BC004004 |
| chr16_-_45664664 | 0.63 |
ENSMUST00000036355.13
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr13_+_93440572 | 0.63 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr10_+_75402090 | 0.62 |
ENSMUST00000129232.8
ENSMUST00000143792.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr1_-_9369463 | 0.62 |
ENSMUST00000140295.8
|
Sntg1
|
syntrophin, gamma 1 |
| chr18_+_22478146 | 0.62 |
ENSMUST00000120223.8
ENSMUST00000097655.4 |
Asxl3
|
ASXL transcriptional regulator 3 |
| chr13_+_27241551 | 0.62 |
ENSMUST00000110369.10
ENSMUST00000224228.2 ENSMUST00000018061.7 |
Prl
|
prolactin |
| chr11_-_98220466 | 0.61 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr9_+_118307250 | 0.61 |
ENSMUST00000111763.8
|
Eomes
|
eomesodermin |
| chr1_+_132119169 | 0.60 |
ENSMUST00000188169.7
ENSMUST00000112357.9 ENSMUST00000188175.2 |
Lemd1
Gm29695
|
LEM domain containing 1 predicted gene, 29695 |
| chr16_+_11224481 | 0.60 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr11_-_99433984 | 0.59 |
ENSMUST00000107443.8
ENSMUST00000074253.4 |
Krt40
|
keratin 40 |
| chr12_-_111947536 | 0.58 |
ENSMUST00000185354.2
|
Rd3l
|
retinal degeneration 3-like |
| chr16_+_17712061 | 0.58 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr9_+_118892497 | 0.57 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chr10_+_112151245 | 0.57 |
ENSMUST00000218445.2
|
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr9_+_123921573 | 0.57 |
ENSMUST00000111442.3
ENSMUST00000171499.3 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr14_+_53310220 | 0.57 |
ENSMUST00000196079.2
|
Trav13d-4
|
T cell receptor alpha variable 13D-4 |
| chr2_+_132689640 | 0.57 |
ENSMUST00000124836.8
ENSMUST00000154160.2 |
Crls1
|
cardiolipin synthase 1 |
| chr7_-_37472290 | 0.57 |
ENSMUST00000176205.8
|
Zfp536
|
zinc finger protein 536 |
| chr3_+_18108313 | 0.57 |
ENSMUST00000026120.8
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr11_-_99519057 | 0.56 |
ENSMUST00000081007.7
|
Krtap4-1
|
keratin associated protein 4-1 |
| chr14_-_96756503 | 0.56 |
ENSMUST00000022666.9
|
Klhl1
|
kelch-like 1 |
| chr13_+_67980849 | 0.56 |
ENSMUST00000221620.2
|
Gm10037
|
predicted gene 10037 |
| chr5_+_104318542 | 0.55 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr6_-_29507944 | 0.55 |
ENSMUST00000101614.10
ENSMUST00000078112.13 |
Kcp
|
kielin/chordin-like protein |
| chr3_-_75072319 | 0.54 |
ENSMUST00000124618.2
|
Zbbx
|
zinc finger, B-box domain containing |
| chr17_-_37399343 | 0.54 |
ENSMUST00000207101.2
ENSMUST00000217397.2 ENSMUST00000215195.2 ENSMUST00000216488.2 |
Olfr90
|
olfactory receptor 90 |
| chr14_+_47710574 | 0.53 |
ENSMUST00000228740.2
|
Fbxo34
|
F-box protein 34 |
| chr5_+_118307754 | 0.53 |
ENSMUST00000054836.7
|
Hrk
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr8_+_84262409 | 0.53 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr14_-_63221950 | 0.52 |
ENSMUST00000100493.3
|
Defb48
|
defensin beta 48 |
| chr11_+_73489420 | 0.51 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr12_+_59060162 | 0.50 |
ENSMUST00000021379.8
|
Gemin2
|
gem nuclear organelle associated protein 2 |
| chr2_+_87686206 | 0.50 |
ENSMUST00000217376.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr7_-_141710850 | 0.50 |
ENSMUST00000209599.2
|
Gm29735
|
predicted gene, 29735 |
| chr4_+_135870808 | 0.50 |
ENSMUST00000008016.3
|
Id3
|
inhibitor of DNA binding 3 |
| chr11_-_49004584 | 0.49 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr11_+_31823096 | 0.49 |
ENSMUST00000155278.2
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_-_110983281 | 0.49 |
ENSMUST00000132464.2
|
Gm15130
|
predicted gene 15130 |
| chr7_-_24211879 | 0.49 |
ENSMUST00000234781.2
|
Gm50092
|
predicted gene, 50092 |
| chr3_+_18002574 | 0.49 |
ENSMUST00000029080.5
|
Cypt12
|
cysteine-rich perinuclear theca 12 |
| chr3_+_132335704 | 0.48 |
ENSMUST00000212594.2
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr3_-_59127571 | 0.47 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr7_-_102540089 | 0.47 |
ENSMUST00000217024.2
|
Olfr569
|
olfactory receptor 569 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 2.1 | 8.5 | GO:0010269 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 1.8 | 5.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 1.3 | 3.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.9 | 4.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.6 | 3.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.5 | 3.2 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.5 | 2.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.5 | 1.5 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.4 | 1.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.4 | 6.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 1.5 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.3 | 6.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.3 | 1.3 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.3 | 10.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 3.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 3.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 1.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 | 5.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 3.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.2 | 2.8 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 2.6 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.8 | GO:0032329 | serine transport(GO:0032329) |
| 0.2 | 2.9 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 0.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.2 | 0.6 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 7.3 | GO:0097286 | iron ion import(GO:0097286) |
| 0.2 | 1.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 0.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 | 0.3 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.2 | 1.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.8 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 2.5 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 0.8 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.2 | 1.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 1.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 1.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.8 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.8 | GO:0051611 | regulation of serotonin uptake(GO:0051611) |
| 0.1 | 1.3 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.5 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 2.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.6 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 2.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.9 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 2.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 3.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 1.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.5 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 5.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.8 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 0.7 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.7 | GO:0097106 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) postsynaptic density organization(GO:0097106) |
| 0.1 | 0.6 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 1.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 2.0 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 0.5 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.8 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 4.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.6 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.1 | 0.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.2 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 | 0.3 | GO:0032053 | ciliary basal body organization(GO:0032053) cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 0.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.3 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.3 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.9 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 1.0 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.3 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 1.0 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.4 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.8 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.4 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 2.3 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.9 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 1.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.6 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.7 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.3 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 2.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.8 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 0.9 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 1.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.4 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 0.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.8 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.3 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 2.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.6 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 2.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.2 | GO:0050718 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.8 | 4.7 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 1.3 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.3 | 2.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 1.8 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 4.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 0.9 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 1.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 2.0 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 1.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 2.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 6.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 3.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 5.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 5.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 3.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 13.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 6.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 2.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.0 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.6 | 6.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.6 | 7.8 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 1.3 | 4.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.2 | 3.6 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.8 | 3.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.8 | 5.5 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.5 | 3.7 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 1.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.3 | 2.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 1.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.3 | 1.0 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.3 | 4.7 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.3 | 10.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 2.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 5.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 8.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 0.7 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.2 | 0.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 3.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 2.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 6.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 2.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 2.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 5.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 3.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 3.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 2.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 2.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 1.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 5.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.3 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 4.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 6.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 4.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 8.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 2.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 6.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 4.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 2.3 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 0.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 3.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 3.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 3.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |