Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
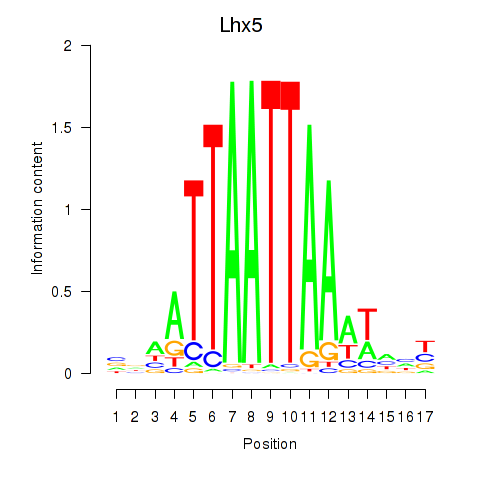
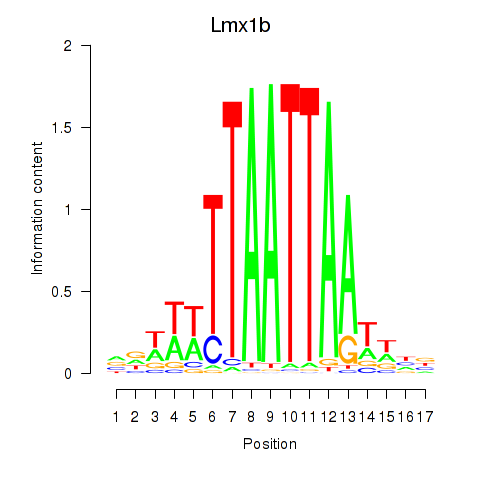
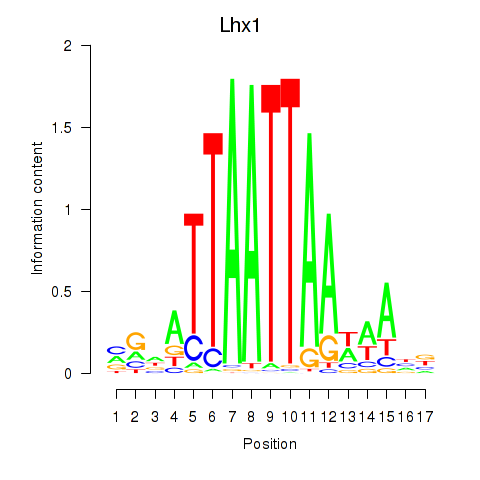
Results for Lhx5_Lmx1b_Lhx1
Z-value: 0.82
Transcription factors associated with Lhx5_Lmx1b_Lhx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx5
|
ENSMUSG00000029595.10 | Lhx5 |
|
Lmx1b
|
ENSMUSG00000038765.15 | Lmx1b |
|
Lhx1
|
ENSMUSG00000018698.16 | Lhx1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx5 | mm39_v1_chr5_+_120569764_120569764 | 0.25 | 3.1e-02 | Click! |
| Lmx1b | mm39_v1_chr2_-_33530492_33530620 | 0.14 | 2.3e-01 | Click! |
| Lhx1 | mm39_v1_chr11_-_84416340_84416369 | 0.08 | 4.9e-01 | Click! |
Activity profile of Lhx5_Lmx1b_Lhx1 motif
Sorted Z-values of Lhx5_Lmx1b_Lhx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx5_Lmx1b_Lhx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_118382926 | 8.73 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr13_+_46655589 | 7.55 |
ENSMUST00000119341.2
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr13_+_46655324 | 6.56 |
ENSMUST00000021802.16
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr13_+_46655617 | 6.17 |
ENSMUST00000225824.2
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr17_+_70828649 | 5.99 |
ENSMUST00000233283.2
|
Dlgap1
|
DLG associated protein 1 |
| chr18_-_38999755 | 5.88 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr2_+_124978518 | 5.84 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr3_+_7494108 | 5.81 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr18_-_39000056 | 5.67 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr1_-_158183894 | 5.53 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr6_+_145879839 | 5.37 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr10_+_29074950 | 5.25 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr19_-_41065544 | 5.09 |
ENSMUST00000087176.8
|
Opalin
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr2_+_124978612 | 5.08 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr13_+_67981349 | 4.72 |
ENSMUST00000222626.2
ENSMUST00000060609.8 |
Gm10037
|
predicted gene 10037 |
| chr9_-_14292453 | 4.30 |
ENSMUST00000167549.2
|
Endod1
|
endonuclease domain containing 1 |
| chr8_-_95929420 | 3.75 |
ENSMUST00000212842.2
|
Kifc3
|
kinesin family member C3 |
| chrX_+_16485937 | 3.52 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr15_+_16778187 | 3.50 |
ENSMUST00000026432.8
|
Cdh9
|
cadherin 9 |
| chr9_+_113641615 | 3.48 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr10_+_127226180 | 3.46 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chrX_+_93679671 | 3.06 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr11_+_59197746 | 2.97 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr15_-_93493758 | 2.95 |
ENSMUST00000048982.11
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr14_+_69585036 | 2.93 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr5_+_13448833 | 2.83 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chrX_+_158242121 | 2.80 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr1_+_66403774 | 2.77 |
ENSMUST00000077355.12
ENSMUST00000114012.8 |
Map2
|
microtubule-associated protein 2 |
| chr11_+_67689094 | 2.71 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr18_-_73887528 | 2.69 |
ENSMUST00000041138.3
|
Elac1
|
elaC ribonuclease Z 1 |
| chr3_+_55369149 | 2.64 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr5_+_104350475 | 2.61 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr18_-_64649620 | 2.55 |
ENSMUST00000025483.11
ENSMUST00000237400.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr1_-_65162267 | 2.49 |
ENSMUST00000050047.4
ENSMUST00000148020.8 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr7_+_54485336 | 2.45 |
ENSMUST00000082373.8
|
Luzp2
|
leucine zipper protein 2 |
| chr14_+_65504067 | 2.41 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr2_+_83642910 | 2.39 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr13_+_67981973 | 2.30 |
ENSMUST00000231606.2
|
Gm10037
|
predicted gene 10037 |
| chrX_+_151922936 | 2.29 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chr10_+_115979787 | 2.28 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr8_-_68363564 | 2.27 |
ENSMUST00000093468.12
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_+_130398261 | 2.19 |
ENSMUST00000086029.10
|
Caln1
|
calneuron 1 |
| chr13_+_77283632 | 2.14 |
ENSMUST00000168779.3
ENSMUST00000225605.2 |
2210408I21Rik
|
RIKEN cDNA 2210408I21 gene |
| chr11_-_74238498 | 2.10 |
ENSMUST00000080365.6
|
Olfr411
|
olfactory receptor 411 |
| chr18_-_15536747 | 2.08 |
ENSMUST00000079081.8
|
Aqp4
|
aquaporin 4 |
| chr1_+_17215581 | 2.07 |
ENSMUST00000026879.8
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1 |
| chr6_+_8948608 | 2.06 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr11_+_109434519 | 2.05 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chrX_-_74621828 | 1.98 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr17_+_70829050 | 1.97 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr11_-_11920540 | 1.89 |
ENSMUST00000109653.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chrX_+_9751861 | 1.89 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr2_+_67935015 | 1.87 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr9_-_54381409 | 1.86 |
ENSMUST00000127880.8
|
Dmxl2
|
Dmx-like 2 |
| chr1_+_109911467 | 1.84 |
ENSMUST00000172005.8
|
Cdh7
|
cadherin 7, type 2 |
| chr14_+_7030760 | 1.79 |
ENSMUST00000055211.6
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chrX_-_142716200 | 1.78 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr4_+_147576874 | 1.75 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982 |
| chr5_+_9316097 | 1.75 |
ENSMUST00000134991.8
ENSMUST00000069538.14 ENSMUST00000115348.9 |
Elapor2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr10_+_23672842 | 1.71 |
ENSMUST00000119597.8
ENSMUST00000179321.8 ENSMUST00000133289.2 |
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr11_-_87249837 | 1.69 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr2_-_29677634 | 1.68 |
ENSMUST00000177467.8
ENSMUST00000113807.10 |
Trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr8_+_94537910 | 1.65 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr2_+_65451100 | 1.64 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr12_-_84664001 | 1.62 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr18_-_66993567 | 1.60 |
ENSMUST00000057942.4
|
Mc4r
|
melanocortin 4 receptor |
| chr10_-_20424069 | 1.60 |
ENSMUST00000169404.8
|
Pde7b
|
phosphodiesterase 7B |
| chr18_+_86729184 | 1.58 |
ENSMUST00000068423.10
|
Cbln2
|
cerebellin 2 precursor protein |
| chr9_-_49710190 | 1.58 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr16_+_11224481 | 1.57 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr18_+_57601541 | 1.56 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr2_+_20742115 | 1.53 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr10_-_20424101 | 1.51 |
ENSMUST00000164195.2
|
Pde7b
|
phosphodiesterase 7B |
| chr14_-_104081119 | 1.50 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr17_-_40630096 | 1.49 |
ENSMUST00000026498.5
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chr18_+_37847792 | 1.48 |
ENSMUST00000192511.2
ENSMUST00000193476.2 |
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr9_-_49710058 | 1.46 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr18_+_37888770 | 1.45 |
ENSMUST00000061279.10
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr18_+_31742565 | 1.45 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr16_+_58967409 | 1.44 |
ENSMUST00000216957.3
|
Olfr195
|
olfactory receptor 195 |
| chr11_-_4045343 | 1.43 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr7_-_10011933 | 1.40 |
ENSMUST00000227719.2
ENSMUST00000228622.2 ENSMUST00000228086.2 |
Vmn1r66
|
vomeronasal 1 receptor 66 |
| chr11_+_61896161 | 1.39 |
ENSMUST00000201624.4
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr18_+_37100672 | 1.38 |
ENSMUST00000193777.6
ENSMUST00000193389.2 |
Pcdha6
|
protocadherin alpha 6 |
| chr9_+_57411279 | 1.37 |
ENSMUST00000080514.9
|
Rpp25
|
ribonuclease P/MRP 25 subunit |
| chrX_-_149440362 | 1.37 |
ENSMUST00000148604.2
|
Tro
|
trophinin |
| chr18_+_38429792 | 1.36 |
ENSMUST00000237211.2
|
Rnf14
|
ring finger protein 14 |
| chr10_+_129493563 | 1.36 |
ENSMUST00000217094.2
|
Olfr800
|
olfactory receptor 800 |
| chr1_-_63215812 | 1.34 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr18_+_37153149 | 1.33 |
ENSMUST00000047614.4
|
Pcdha12
|
protocadherin alpha 12 |
| chr2_-_20948230 | 1.29 |
ENSMUST00000140230.2
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr3_-_72875187 | 1.28 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr6_-_113478779 | 1.27 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr2_-_73410632 | 1.25 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr18_+_62681982 | 1.22 |
ENSMUST00000055725.12
ENSMUST00000162365.8 |
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr1_-_135513083 | 1.21 |
ENSMUST00000040599.15
|
Nav1
|
neuron navigator 1 |
| chr2_-_86944911 | 1.20 |
ENSMUST00000216088.3
|
Olfr259
|
olfactory receptor 259 |
| chr1_-_63215952 | 1.18 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr2_+_32496990 | 1.18 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr7_-_19297001 | 1.16 |
ENSMUST00000058444.10
|
Ppp1r37
|
protein phosphatase 1, regulatory subunit 37 |
| chr8_-_65582206 | 1.15 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr14_-_104081827 | 1.15 |
ENSMUST00000022718.11
|
Ednrb
|
endothelin receptor type B |
| chr7_-_142209755 | 1.15 |
ENSMUST00000178921.2
|
Igf2
|
insulin-like growth factor 2 |
| chr8_-_123087485 | 1.14 |
ENSMUST00000044123.2
|
Trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr13_+_23433408 | 1.14 |
ENSMUST00000091719.2
|
Vmn1r223
|
vomeronasal 1 receptor 223 |
| chr12_+_10440755 | 1.14 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr10_+_34173426 | 1.12 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr6_-_41423004 | 1.08 |
ENSMUST00000095999.7
|
Gm10334
|
predicted gene 10334 |
| chr9_-_15212745 | 1.08 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chrM_+_14138 | 1.07 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr17_+_29493157 | 1.05 |
ENSMUST00000234234.2
|
BC004004
|
cDNA sequence BC004004 |
| chr16_-_29363671 | 1.04 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr1_-_83016152 | 1.04 |
ENSMUST00000164473.2
ENSMUST00000045560.15 |
Slc19a3
|
solute carrier family 19, member 3 |
| chr1_+_179928709 | 1.04 |
ENSMUST00000133890.8
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_-_5068743 | 1.03 |
ENSMUST00000027986.5
|
Optn
|
optineurin |
| chr3_-_66204228 | 1.01 |
ENSMUST00000029419.8
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr9_+_80072361 | 1.01 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr11_+_51152546 | 1.01 |
ENSMUST00000130641.8
|
Clk4
|
CDC like kinase 4 |
| chr1_+_167445815 | 0.99 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr16_-_45352346 | 0.99 |
ENSMUST00000232600.2
|
Gm17783
|
predicted gene, 17783 |
| chr18_-_64649497 | 0.98 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chrM_-_14061 | 0.97 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr8_+_21515561 | 0.96 |
ENSMUST00000076754.3
|
Defa21
|
defensin, alpha, 21 |
| chr11_-_80268800 | 0.95 |
ENSMUST00000179332.8
ENSMUST00000103225.11 ENSMUST00000134274.2 |
5730455P16Rik
|
RIKEN cDNA 5730455P16 gene |
| chr11_-_109886601 | 0.95 |
ENSMUST00000020948.15
|
Abca8b
|
ATP-binding cassette, sub-family A (ABC1), member 8b |
| chr8_-_49008305 | 0.95 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr2_-_5068807 | 0.94 |
ENSMUST00000114996.8
|
Optn
|
optineurin |
| chr10_+_94411119 | 0.94 |
ENSMUST00000121471.8
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr10_+_90412114 | 0.93 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr18_+_38430205 | 0.93 |
ENSMUST00000235491.2
|
Rnf14
|
ring finger protein 14 |
| chr1_-_174749379 | 0.91 |
ENSMUST00000055294.4
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr18_+_37125424 | 0.91 |
ENSMUST00000194038.2
|
Pcdha8
|
protocadherin alpha 8 |
| chr17_+_79919267 | 0.90 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr9_-_117843228 | 0.89 |
ENSMUST00000187803.3
|
Zcwpw2
|
zinc finger, CW type with PWWP domain 2 |
| chr11_-_109886569 | 0.88 |
ENSMUST00000106669.3
|
Abca8b
|
ATP-binding cassette, sub-family A (ABC1), member 8b |
| chr18_+_23885390 | 0.86 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chrX_+_168468186 | 0.86 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr18_+_38429858 | 0.85 |
ENSMUST00000171461.3
ENSMUST00000235811.2 |
Rnf14
|
ring finger protein 14 |
| chr6_+_134617903 | 0.84 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr9_+_110948492 | 0.84 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr10_-_129965752 | 0.84 |
ENSMUST00000215217.2
ENSMUST00000214192.2 |
Olfr824
|
olfactory receptor 824 |
| chr2_+_85606930 | 0.83 |
ENSMUST00000076250.2
|
Olfr1014
|
olfactory receptor 1014 |
| chrX_+_100492684 | 0.82 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr9_+_38738911 | 0.82 |
ENSMUST00000051238.7
ENSMUST00000219798.2 |
Olfr923
|
olfactory receptor 923 |
| chr13_+_67980849 | 0.81 |
ENSMUST00000221620.2
|
Gm10037
|
predicted gene 10037 |
| chr8_+_21691577 | 0.80 |
ENSMUST00000110754.2
|
Defa41
|
defensin, alpha, 41 |
| chr11_+_105956867 | 0.80 |
ENSMUST00000002048.8
|
Taco1
|
translational activator of mitochondrially encoded cytochrome c oxidase I |
| chr7_-_5325456 | 0.79 |
ENSMUST00000207520.2
|
Nlrp2
|
NLR family, pyrin domain containing 2 |
| chr9_+_92339422 | 0.78 |
ENSMUST00000034941.9
|
Plscr4
|
phospholipid scramblase 4 |
| chr16_-_44153498 | 0.78 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr10_-_75946790 | 0.77 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr16_-_44153288 | 0.77 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr2_-_89972637 | 0.77 |
ENSMUST00000215659.2
ENSMUST00000215765.2 ENSMUST00000213293.2 ENSMUST00000214973.2 ENSMUST00000215153.3 |
Olfr32
|
olfactory receptor 32 |
| chr8_+_107757847 | 0.76 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr8_-_27903965 | 0.76 |
ENSMUST00000033882.10
|
Chrna6
|
cholinergic receptor, nicotinic, alpha polypeptide 6 |
| chr13_-_4659120 | 0.75 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr14_+_26616514 | 0.74 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr9_-_79920131 | 0.74 |
ENSMUST00000217264.2
|
Filip1
|
filamin A interacting protein 1 |
| chr19_+_5524701 | 0.74 |
ENSMUST00000165485.8
ENSMUST00000166253.8 ENSMUST00000167371.8 ENSMUST00000167855.8 ENSMUST00000070118.14 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr7_+_99808526 | 0.74 |
ENSMUST00000207825.2
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chrX_+_156482116 | 0.74 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr4_+_108576846 | 0.73 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr2_-_120946877 | 0.73 |
ENSMUST00000110675.3
|
Tgm7
|
transglutaminase 7 |
| chr2_-_85966272 | 0.73 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr4_+_102278715 | 0.72 |
ENSMUST00000106904.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr12_+_88327305 | 0.72 |
ENSMUST00000101165.9
|
Adck1
|
aarF domain containing kinase 1 |
| chr8_+_121842902 | 0.71 |
ENSMUST00000054691.8
|
Foxc2
|
forkhead box C2 |
| chr5_-_3697806 | 0.71 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr17_+_29493113 | 0.70 |
ENSMUST00000234326.2
ENSMUST00000235117.2 |
BC004004
|
cDNA sequence BC004004 |
| chr10_-_23112973 | 0.70 |
ENSMUST00000218049.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr10_-_56104732 | 0.68 |
ENSMUST00000099739.5
|
Tbc1d32
|
TBC1 domain family, member 32 |
| chr7_+_99808452 | 0.67 |
ENSMUST00000032967.4
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr18_+_38430015 | 0.66 |
ENSMUST00000236319.2
|
Rnf14
|
ring finger protein 14 |
| chr14_-_57809008 | 0.65 |
ENSMUST00000022518.8
ENSMUST00000239099.2 |
Eef1akmt1
|
EEF1A alpha lysine methyltransferase 1 |
| chr4_+_145237329 | 0.65 |
ENSMUST00000105742.8
ENSMUST00000136309.8 |
Zfp990
|
zinc finger protein 990 |
| chr2_-_101713355 | 0.65 |
ENSMUST00000154525.2
|
Prr5l
|
proline rich 5 like |
| chr2_+_163500290 | 0.64 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr2_-_36975758 | 0.63 |
ENSMUST00000100145.2
|
Olfr361
|
olfactory receptor 361 |
| chr18_+_37898633 | 0.62 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr5_-_108943211 | 0.61 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr14_+_67470884 | 0.60 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr7_-_12340471 | 0.60 |
ENSMUST00000170412.2
|
Vmn2r53
|
vomeronasal 2, receptor 53 |
| chr10_+_97318223 | 0.60 |
ENSMUST00000163448.4
|
Dcn
|
decorin |
| chr19_-_12313274 | 0.60 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr12_+_88327391 | 0.60 |
ENSMUST00000222695.2
|
Adck1
|
aarF domain containing kinase 1 |
| chr9_-_50650663 | 0.60 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr11_-_49004584 | 0.59 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr8_+_65399831 | 0.59 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr3_-_113371392 | 0.59 |
ENSMUST00000067980.12
|
Amy1
|
amylase 1, salivary |
| chr7_+_19927635 | 0.57 |
ENSMUST00000168984.2
|
Vmn1r95
|
vomeronasal 1 receptor, 95 |
| chrX_+_73352694 | 0.56 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr15_+_99192968 | 0.55 |
ENSMUST00000128352.8
ENSMUST00000145482.8 |
Prpf40b
|
pre-mRNA processing factor 40B |
| chr5_-_129747129 | 0.55 |
ENSMUST00000049778.7
|
Zfp11
|
zinc finger protein 11 |
| chr6_-_50374048 | 0.55 |
ENSMUST00000136926.3
|
Osbpl3
|
oxysterol binding protein-like 3 |
| chr2_+_69727563 | 0.55 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr17_+_29493049 | 0.54 |
ENSMUST00000149405.4
|
BC004004
|
cDNA sequence BC004004 |
| chr7_+_43077088 | 0.54 |
ENSMUST00000239023.2
|
Gm38999
|
predicted gene, 38999 |
| chr10_-_129107354 | 0.53 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr19_-_33764859 | 0.53 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr2_-_174188505 | 0.53 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.5 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 1.0 | 3.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 1.0 | 2.9 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.9 | 2.7 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.8 | 3.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.7 | 3.7 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.6 | 8.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.5 | 3.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.5 | 2.6 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.5 | 3.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.5 | 1.4 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.4 | 2.7 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.4 | 2.7 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.4 | 2.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.4 | 2.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.3 | 2.0 | GO:0061734 | negative regulation of receptor recycling(GO:0001920) parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.3 | 3.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 6.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 0.9 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.3 | 1.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 0.8 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.3 | 1.4 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.3 | 1.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 0.5 | GO:2000722 | regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.3 | 0.8 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.2 | 20.3 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.2 | 2.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 1.9 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.0 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.2 | 2.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 1.9 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 0.7 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 0.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 1.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.7 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 1.6 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 1.9 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 2.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.5 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.4 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 1.8 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 1.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.4 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 3.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 1.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.3 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.1 | 0.4 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 1.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.9 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 3.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 3.8 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.1 | 2.7 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.1 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 5.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 1.2 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 1.0 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 2.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.2 | GO:0036343 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.1 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.8 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.8 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 2.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.7 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.2 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 1.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 1.0 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 1.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 4.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:0051611 | regulation of serotonin uptake(GO:0051611) |
| 0.0 | 2.6 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.8 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 1.5 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 1.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 3.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.4 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 20.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.4 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 1.8 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 1.4 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.3 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.6 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.8 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 1.1 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 1.0 | 3.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.9 | 2.7 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.7 | 2.9 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.6 | 1.9 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.5 | 2.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 2.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 3.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 1.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 5.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 19.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.8 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 2.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.1 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 2.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 5.5 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 2.3 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 2.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 14.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 10.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 3.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 5.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 8.6 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 4.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.9 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.9 | 3.5 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.8 | 20.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 1.6 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.5 | 11.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 1.9 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.4 | 3.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 3.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.4 | 2.9 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.3 | 1.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 2.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 6.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 0.8 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.3 | 1.6 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 1.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 2.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 2.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 1.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 0.5 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.2 | 2.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 1.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.8 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 4.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 4.1 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 0.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.0 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 2.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.3 | GO:0031692 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 2.0 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 2.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.9 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 4.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.3 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.1 | 0.2 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 2.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 3.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.6 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 3.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 2.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 2.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 13.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 2.5 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 3.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 2.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 11.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 4.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 1.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 14.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 2.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 2.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 4.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 5.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.4 | 20.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 3.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 4.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 4.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 2.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |