Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Maf_Nrl
Z-value: 1.12
Transcription factors associated with Maf_Nrl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
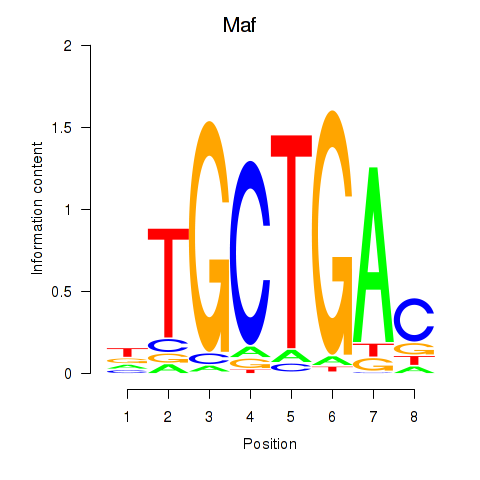
Maf
|
ENSMUSG00000055435.7 | Maf |
|
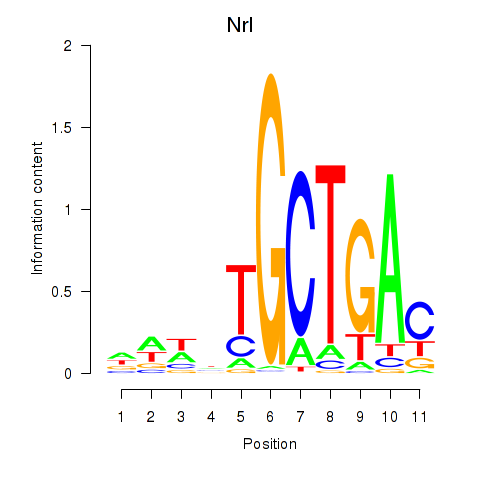
Nrl
|
ENSMUSG00000040632.17 | Nrl |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Maf | mm39_v1_chr8_-_116433733_116433835 | 0.37 | 1.4e-03 | Click! |
| Nrl | mm39_v1_chr14_-_55762416_55762423 | 0.25 | 3.4e-02 | Click! |
Activity profile of Maf_Nrl motif
Sorted Z-values of Maf_Nrl motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Maf_Nrl
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_66974492 | 14.00 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr1_-_66974694 | 13.11 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr2_+_120307390 | 6.39 |
ENSMUST00000110716.9
ENSMUST00000028748.14 ENSMUST00000090028.13 ENSMUST00000110719.4 |
Capn3
|
calpain 3 |
| chr4_+_104623505 | 6.36 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr10_+_87695886 | 5.54 |
ENSMUST00000062862.13
|
Igf1
|
insulin-like growth factor 1 |
| chr10_+_87695352 | 5.53 |
ENSMUST00000121952.8
ENSMUST00000126490.8 |
Igf1
|
insulin-like growth factor 1 |
| chr1_-_66984178 | 4.85 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr2_-_76812799 | 4.82 |
ENSMUST00000011934.13
ENSMUST00000099981.10 ENSMUST00000099980.10 ENSMUST00000111882.9 ENSMUST00000140091.8 |
Ttn
|
titin |
| chr1_-_66984521 | 4.63 |
ENSMUST00000160100.2
|
Myl1
|
myosin, light polypeptide 1 |
| chr11_+_67167950 | 4.61 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr7_-_126062272 | 4.00 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr11_-_78950641 | 3.99 |
ENSMUST00000226282.2
|
Ksr1
|
kinase suppressor of ras 1 |
| chr9_-_46146558 | 3.92 |
ENSMUST00000121916.8
ENSMUST00000034586.9 |
Apoc3
|
apolipoprotein C-III |
| chr11_-_53313950 | 3.82 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr5_+_110692162 | 3.54 |
ENSMUST00000040001.14
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr16_+_17149235 | 3.51 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr14_-_21764638 | 3.23 |
ENSMUST00000073870.7
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr11_-_78950698 | 3.23 |
ENSMUST00000141409.8
|
Ksr1
|
kinase suppressor of ras 1 |
| chr6_-_124441731 | 3.05 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chrX_+_149377416 | 3.01 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chrX_+_41591355 | 2.95 |
ENSMUST00000189753.7
|
Sh2d1a
|
SH2 domain containing 1A |
| chr5_+_31274064 | 2.86 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr4_+_143076327 | 2.68 |
ENSMUST00000052458.3
|
Lrrc38
|
leucine rich repeat containing 38 |
| chr13_+_83652352 | 2.62 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr10_+_87695117 | 2.62 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr3_+_99203818 | 2.54 |
ENSMUST00000150756.3
|
Tbx15
|
T-box 15 |
| chr10_+_112292161 | 2.51 |
ENSMUST00000219607.2
ENSMUST00000218827.2 |
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr13_+_83720457 | 2.46 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr5_+_31274046 | 2.45 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr16_+_20514925 | 2.44 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr2_-_42543069 | 2.44 |
ENSMUST00000203080.3
|
Lrp1b
|
low density lipoprotein-related protein 1B |
| chr9_-_46146928 | 2.35 |
ENSMUST00000118649.8
|
Apoc3
|
apolipoprotein C-III |
| chr14_-_34310637 | 2.30 |
ENSMUST00000227819.2
|
Ldb3
|
LIM domain binding 3 |
| chr4_-_63072367 | 2.29 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr10_+_87696339 | 2.28 |
ENSMUST00000121161.8
|
Igf1
|
insulin-like growth factor 1 |
| chr16_+_20513658 | 2.28 |
ENSMUST00000056518.13
|
Fam131a
|
family with sequence similarity 131, member A |
| chr3_-_33039225 | 2.19 |
ENSMUST00000108221.2
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr13_+_83652150 | 2.19 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr2_-_151474391 | 2.15 |
ENSMUST00000137936.2
ENSMUST00000146172.8 ENSMUST00000094456.10 ENSMUST00000148755.8 ENSMUST00000109875.8 ENSMUST00000028951.14 ENSMUST00000109877.10 |
Snph
|
syntaphilin |
| chr13_+_83652280 | 2.13 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr13_+_83720484 | 2.12 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr4_-_6275629 | 2.10 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr1_-_163552693 | 2.09 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr3_-_33137209 | 2.06 |
ENSMUST00000194016.6
ENSMUST00000193681.6 ENSMUST00000192093.6 ENSMUST00000193289.6 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr15_-_41733099 | 2.06 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein |
| chr3_-_95135946 | 2.06 |
ENSMUST00000065482.6
|
Mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr2_+_51928017 | 2.05 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr7_-_19411866 | 2.00 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr3_-_33137165 | 1.93 |
ENSMUST00000078226.10
ENSMUST00000108224.8 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr15_+_89338116 | 1.91 |
ENSMUST00000023291.6
|
Mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr11_-_106240215 | 1.89 |
ENSMUST00000021056.8
|
Scn4a
|
sodium channel, voltage-gated, type IV, alpha |
| chr9_+_110074574 | 1.87 |
ENSMUST00000197850.5
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr10_+_4660119 | 1.84 |
ENSMUST00000105588.8
ENSMUST00000105589.2 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr7_-_74958121 | 1.83 |
ENSMUST00000085164.7
|
Sv2b
|
synaptic vesicle glycoprotein 2 b |
| chr10_+_87694924 | 1.83 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr9_+_44309727 | 1.83 |
ENSMUST00000213268.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr14_-_34310438 | 1.78 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr3_+_45332831 | 1.71 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr5_+_112403678 | 1.68 |
ENSMUST00000031286.13
ENSMUST00000131673.8 ENSMUST00000112375.2 |
Crybb1
|
crystallin, beta B1 |
| chr15_-_66985760 | 1.65 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr12_-_90705212 | 1.64 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr14_-_34310602 | 1.62 |
ENSMUST00000064098.14
ENSMUST00000090040.12 ENSMUST00000022330.9 ENSMUST00000022327.13 |
Ldb3
|
LIM domain binding 3 |
| chr5_+_138340203 | 1.60 |
ENSMUST00000211088.2
|
Pvrig
|
poliovirus receptor related immunoglobulin domain containing |
| chr18_-_43192483 | 1.57 |
ENSMUST00000025377.14
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_-_167030706 | 1.56 |
ENSMUST00000207917.2
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr11_+_54195006 | 1.54 |
ENSMUST00000108904.10
ENSMUST00000108905.10 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr3_-_158267771 | 1.49 |
ENSMUST00000199890.5
ENSMUST00000238317.3 ENSMUST00000200137.5 ENSMUST00000106044.6 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr2_-_118203826 | 1.47 |
ENSMUST00000039160.3
|
Gpr176
|
G protein-coupled receptor 176 |
| chr7_-_79036691 | 1.47 |
ENSMUST00000053718.15
ENSMUST00000179243.3 |
Rlbp1
|
retinaldehyde binding protein 1 |
| chr2_-_27032441 | 1.47 |
ENSMUST00000151224.3
|
Fam163b
|
family with sequence similarity 163, member B |
| chr7_-_79036222 | 1.44 |
ENSMUST00000205638.2
ENSMUST00000206320.3 ENSMUST00000205442.2 |
Rlbp1
|
retinaldehyde binding protein 1 |
| chr17_+_69746321 | 1.44 |
ENSMUST00000169935.2
|
Akain1
|
A kinase (PRKA) anchor inhibitor 1 |
| chr11_+_97732108 | 1.43 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr9_+_46139878 | 1.37 |
ENSMUST00000034588.9
ENSMUST00000132155.2 |
Apoa1
|
apolipoprotein A-I |
| chr19_+_38252984 | 1.35 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr19_-_45224251 | 1.32 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chrX_+_41591476 | 1.31 |
ENSMUST00000115070.8
ENSMUST00000153948.2 |
Sh2d1a
|
SH2 domain containing 1A |
| chr16_+_39804711 | 1.30 |
ENSMUST00000187695.7
|
Lsamp
|
limbic system-associated membrane protein |
| chr3_-_137773149 | 1.30 |
ENSMUST00000053318.4
|
Gm5105
|
predicted gene 5105 |
| chr10_+_79716876 | 1.30 |
ENSMUST00000166201.2
|
Prtn3
|
proteinase 3 |
| chr9_-_57513510 | 1.30 |
ENSMUST00000215487.2
ENSMUST00000045068.10 |
Cplx3
|
complexin 3 |
| chr8_-_3928495 | 1.28 |
ENSMUST00000209176.2
ENSMUST00000011445.8 |
Cd209d
|
CD209d antigen |
| chr16_-_36810810 | 1.27 |
ENSMUST00000075869.13
|
Fbxo40
|
F-box protein 40 |
| chr11_+_54194831 | 1.26 |
ENSMUST00000000145.12
ENSMUST00000138515.8 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chrX_-_50770580 | 1.26 |
ENSMUST00000088172.12
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chrX_+_41591410 | 1.25 |
ENSMUST00000005839.11
|
Sh2d1a
|
SH2 domain containing 1A |
| chr2_-_66271097 | 1.24 |
ENSMUST00000112371.9
ENSMUST00000138910.3 |
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr18_+_5593566 | 1.23 |
ENSMUST00000160910.2
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr5_-_140612993 | 1.22 |
ENSMUST00000199157.2
|
Ttyh3
|
tweety family member 3 |
| chr4_+_129878627 | 1.22 |
ENSMUST00000120204.8
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr16_-_45975440 | 1.22 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chrX_-_50770733 | 1.20 |
ENSMUST00000114871.2
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr7_+_29008840 | 1.19 |
ENSMUST00000137848.2
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr6_+_51500881 | 1.19 |
ENSMUST00000049152.15
|
Snx10
|
sorting nexin 10 |
| chr15_-_76906832 | 1.18 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr11_+_60913386 | 1.18 |
ENSMUST00000089184.11
|
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr11_+_46701619 | 1.18 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr2_+_96148418 | 1.18 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr19_+_38253077 | 1.17 |
ENSMUST00000198045.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr17_+_46739852 | 1.16 |
ENSMUST00000113465.10
ENSMUST00000024764.12 ENSMUST00000165993.2 |
Crip3
|
cysteine-rich protein 3 |
| chrX_-_142716200 | 1.14 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr5_-_113957362 | 1.13 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr16_+_35803794 | 1.13 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr11_-_28534260 | 1.12 |
ENSMUST00000093253.10
ENSMUST00000109502.9 ENSMUST00000042534.15 |
Ccdc85a
|
coiled-coil domain containing 85A |
| chr3_-_33136153 | 1.12 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr5_-_113957318 | 1.12 |
ENSMUST00000201194.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr12_+_88920169 | 1.12 |
ENSMUST00000057634.14
|
Nrxn3
|
neurexin III |
| chr2_-_42542938 | 1.12 |
ENSMUST00000204204.2
|
Lrp1b
|
low density lipoprotein-related protein 1B |
| chr9_-_73876182 | 1.11 |
ENSMUST00000184666.8
|
Unc13c
|
unc-13 homolog C |
| chr6_+_41523664 | 1.11 |
ENSMUST00000103299.3
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr14_+_74878280 | 1.10 |
ENSMUST00000036653.5
|
Htr2a
|
5-hydroxytryptamine (serotonin) receptor 2A |
| chr1_+_180396478 | 1.10 |
ENSMUST00000027777.12
|
Parp1
|
poly (ADP-ribose) polymerase family, member 1 |
| chr19_+_38253105 | 1.09 |
ENSMUST00000196090.2
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr2_-_42543012 | 1.09 |
ENSMUST00000142688.2
|
Lrp1b
|
low density lipoprotein-related protein 1B |
| chr17_-_35391583 | 1.09 |
ENSMUST00000173106.2
|
Aif1
|
allograft inflammatory factor 1 |
| chr2_+_90865958 | 1.09 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr12_-_112477536 | 1.08 |
ENSMUST00000066791.7
|
Tmem179
|
transmembrane protein 179 |
| chr8_+_66419809 | 1.07 |
ENSMUST00000072482.13
|
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr19_+_60744385 | 1.06 |
ENSMUST00000088237.6
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chrX_-_103900834 | 1.06 |
ENSMUST00000033575.7
|
Magee2
|
MAGE family member E2 |
| chr11_-_107228382 | 1.06 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr6_+_41515152 | 1.05 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chr3_-_79053182 | 1.05 |
ENSMUST00000118340.7
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_-_75240551 | 1.04 |
ENSMUST00000186178.7
ENSMUST00000189769.7 ENSMUST00000027404.12 |
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr19_+_42244143 | 1.03 |
ENSMUST00000087123.6
|
Golga7b
|
golgi autoantigen, golgin subfamily a, 7B |
| chr11_-_35689711 | 1.01 |
ENSMUST00000160726.4
|
Fbll1
|
fibrillarin-like 1 |
| chr6_-_138399896 | 1.01 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr17_+_34124078 | 1.00 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr7_-_79036654 | 1.00 |
ENSMUST00000206695.2
|
Rlbp1
|
retinaldehyde binding protein 1 |
| chr8_-_72175949 | 0.98 |
ENSMUST00000125092.2
|
Fcho1
|
FCH domain only 1 |
| chr14_-_49763310 | 0.98 |
ENSMUST00000146164.2
ENSMUST00000138884.8 ENSMUST00000074368.11 ENSMUST00000123534.2 |
Slc35f4
|
solute carrier family 35, member F4 |
| chrX_-_156275231 | 0.98 |
ENSMUST00000112529.8
|
Sms
|
spermine synthase |
| chr5_-_110801213 | 0.96 |
ENSMUST00000042147.6
|
Noc4l
|
NOC4 like |
| chr4_+_129878890 | 0.96 |
ENSMUST00000106017.8
ENSMUST00000121049.8 |
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr11_+_54194624 | 0.96 |
ENSMUST00000093106.12
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr11_-_70578744 | 0.96 |
ENSMUST00000108545.9
ENSMUST00000120261.8 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr6_-_120470768 | 0.95 |
ENSMUST00000178687.2
|
Tmem121b
|
transmembrane protein 121B |
| chr1_-_136188611 | 0.93 |
ENSMUST00000086395.7
|
Gpr25
|
G protein-coupled receptor 25 |
| chr2_-_25209107 | 0.92 |
ENSMUST00000114318.10
ENSMUST00000114310.10 ENSMUST00000114308.10 ENSMUST00000114317.10 ENSMUST00000028335.13 ENSMUST00000114314.10 ENSMUST00000114307.8 |
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr5_-_112400375 | 0.92 |
ENSMUST00000112385.8
|
Cryba4
|
crystallin, beta A4 |
| chr17_-_24428351 | 0.91 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr13_+_16189041 | 0.90 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chr6_+_70675416 | 0.89 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr10_-_109669053 | 0.89 |
ENSMUST00000238286.2
|
Nav3
|
neuron navigator 3 |
| chr13_+_47347301 | 0.89 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr6_+_47021979 | 0.89 |
ENSMUST00000150737.2
|
Cntnap2
|
contactin associated protein-like 2 |
| chr2_-_58990967 | 0.88 |
ENSMUST00000226455.2
ENSMUST00000077687.6 |
Ccdc148
|
coiled-coil domain containing 148 |
| chr17_+_34341766 | 0.86 |
ENSMUST00000042121.11
|
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr6_-_68681962 | 0.85 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr10_+_41763423 | 0.84 |
ENSMUST00000041438.7
|
Sesn1
|
sestrin 1 |
| chr14_+_67470884 | 0.84 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr2_-_25209199 | 0.84 |
ENSMUST00000114312.2
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr5_-_125371162 | 0.84 |
ENSMUST00000127148.2
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr9_+_109881083 | 0.83 |
ENSMUST00000164930.8
ENSMUST00000199498.5 |
Map4
|
microtubule-associated protein 4 |
| chr16_-_18440388 | 0.81 |
ENSMUST00000167388.3
|
Gp1bb
|
glycoprotein Ib, beta polypeptide |
| chr14_+_55728519 | 0.81 |
ENSMUST00000076236.7
|
Carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr11_+_33996920 | 0.80 |
ENSMUST00000052413.12
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr17_+_27171834 | 0.80 |
ENSMUST00000231853.2
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr10_+_79746690 | 0.79 |
ENSMUST00000181321.2
|
Gm26602
|
predicted gene, 26602 |
| chr11_-_70578905 | 0.79 |
ENSMUST00000108544.8
|
Camta2
|
calmodulin binding transcription activator 2 |
| chr12_-_113324852 | 0.78 |
ENSMUST00000223179.2
ENSMUST00000103423.3 |
Ighg3
|
Immunoglobulin heavy constant gamma 3 |
| chr8_-_3976808 | 0.78 |
ENSMUST00000188386.2
ENSMUST00000084086.9 ENSMUST00000171635.8 |
Cd209b
|
CD209b antigen |
| chr10_+_128247492 | 0.78 |
ENSMUST00000171342.3
|
Rnf41
|
ring finger protein 41 |
| chr11_-_109364424 | 0.77 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr6_+_110622533 | 0.76 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr12_+_108572015 | 0.76 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr1_-_80736165 | 0.75 |
ENSMUST00000077946.12
|
Dock10
|
dedicator of cytokinesis 10 |
| chr11_-_106192627 | 0.74 |
ENSMUST00000103071.4
|
Gh
|
growth hormone |
| chr12_-_70394272 | 0.74 |
ENSMUST00000221370.2
|
Trim9
|
tripartite motif-containing 9 |
| chr10_+_128247598 | 0.73 |
ENSMUST00000096386.13
|
Rnf41
|
ring finger protein 41 |
| chr4_+_101365052 | 0.71 |
ENSMUST00000038207.12
|
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr15_+_82012114 | 0.71 |
ENSMUST00000089174.6
|
Ccdc134
|
coiled-coil domain containing 134 |
| chr11_-_70578775 | 0.71 |
ENSMUST00000036299.14
ENSMUST00000119120.2 ENSMUST00000100933.10 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr10_-_127147609 | 0.71 |
ENSMUST00000037290.12
ENSMUST00000171564.8 |
Mars1
|
methionine-tRNA synthetase 1 |
| chrX_+_136552469 | 0.70 |
ENSMUST00000075471.4
|
Il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr2_+_92213256 | 0.69 |
ENSMUST00000054316.9
ENSMUST00000111280.3 |
1700029I15Rik
|
RIKEN cDNA 1700029I15 gene |
| chr4_+_59003121 | 0.69 |
ENSMUST00000095070.4
ENSMUST00000174664.2 |
Dnajc25
Gm20503
|
DnaJ heat shock protein family (Hsp40) member C25 predicted gene 20503 |
| chr11_-_58346806 | 0.69 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr6_+_70495224 | 0.68 |
ENSMUST00000103396.2
|
Igkv3-12
|
immunoglobulin kappa variable 3-12 |
| chr11_+_77107006 | 0.68 |
ENSMUST00000156488.8
ENSMUST00000037912.12 |
Ssh2
|
slingshot protein phosphatase 2 |
| chr11_+_80319424 | 0.67 |
ENSMUST00000173938.8
ENSMUST00000017572.14 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr11_-_102360664 | 0.67 |
ENSMUST00000103086.4
|
Itga2b
|
integrin alpha 2b |
| chr18_-_35788240 | 0.65 |
ENSMUST00000097619.2
|
Prob1
|
proline rich basic protein 1 |
| chr4_-_43578823 | 0.65 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2 |
| chr11_+_74510413 | 0.65 |
ENSMUST00000100866.3
|
Ccdc92b
|
coiled-coil domain containing 92B |
| chr18_-_35788255 | 0.65 |
ENSMUST00000190196.5
|
Prob1
|
proline rich basic protein 1 |
| chr4_+_101365144 | 0.64 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr14_+_73790105 | 0.63 |
ENSMUST00000160507.8
ENSMUST00000022706.7 |
Sucla2
|
succinate-Coenzyme A ligase, ADP-forming, beta subunit |
| chr18_-_62313019 | 0.63 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr2_-_181333597 | 0.63 |
ENSMUST00000108778.8
ENSMUST00000165416.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr16_-_18052937 | 0.62 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr2_+_181409075 | 0.62 |
ENSMUST00000108757.9
|
Myt1
|
myelin transcription factor 1 |
| chr2_-_76720097 | 0.62 |
ENSMUST00000148747.8
|
Ttn
|
titin |
| chr2_-_118310116 | 0.61 |
ENSMUST00000110862.2
ENSMUST00000009693.15 |
Srp14
|
signal recognition particle 14 |
| chr9_-_34967081 | 0.61 |
ENSMUST00000215463.2
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr6_-_83031358 | 0.60 |
ENSMUST00000113962.8
ENSMUST00000089645.13 ENSMUST00000113963.8 |
Htra2
|
HtrA serine peptidase 2 |
| chr11_-_107361525 | 0.60 |
ENSMUST00000103064.10
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr11_+_108814007 | 0.59 |
ENSMUST00000106711.2
|
Axin2
|
axin 2 |
| chr1_+_171386752 | 0.59 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr16_-_19060440 | 0.59 |
ENSMUST00000103751.3
|
Iglv3
|
immunoglobulin lambda variable 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 17.8 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.6 | 6.4 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 1.6 | 6.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 1.3 | 4.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 1.1 | 5.4 | GO:0043056 | forward locomotion(GO:0043056) |
| 1.0 | 11.5 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.9 | 7.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.8 | 2.5 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.8 | 3.8 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.6 | 1.8 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.5 | 2.0 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.4 | 2.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.4 | 2.1 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.4 | 1.6 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.4 | 2.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 6.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 1.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.4 | 1.1 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.4 | 1.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.4 | 1.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.3 | 1.4 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.3 | 1.0 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.3 | 1.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.3 | 1.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 1.8 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.3 | 3.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 5.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 0.9 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.3 | 0.8 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.3 | 1.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.3 | 1.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.2 | 0.7 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.2 | 1.9 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 3.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 | 1.4 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.2 | 4.6 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.2 | 1.5 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.2 | 1.8 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 1.6 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 | 0.8 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 1.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 2.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.2 | 1.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 0.9 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.2 | 1.9 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.2 | 0.9 | GO:0002358 | pre-B cell allelic exclusion(GO:0002331) B cell homeostatic proliferation(GO:0002358) |
| 0.2 | 0.6 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) diaphragm contraction(GO:0002086) |
| 0.2 | 0.5 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 0.2 | 0.6 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.2 | 34.4 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 2.1 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 0.4 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.7 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.8 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 5.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 1.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.2 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.6 | GO:0032423 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 2.7 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 0.5 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.1 | 1.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 6.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 1.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 1.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 1.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.5 | GO:0009624 | response to nematode(GO:0009624) |
| 0.1 | 4.1 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 1.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.8 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.1 | 0.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.5 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 2.1 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.3 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 1.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 2.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 1.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.6 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 1.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 2.7 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 1.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 6.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.7 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 1.5 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 1.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 2.1 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 1.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 3.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 1.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.8 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 3.6 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 0.3 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 | 3.0 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 4.8 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 1.6 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.3 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 2.9 | GO:0007596 | blood coagulation(GO:0007596) coagulation(GO:0050817) |
| 0.0 | 1.9 | GO:0050804 | modulation of synaptic transmission(GO:0050804) |
| 0.0 | 2.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 1.7 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.4 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 17.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.0 | 3.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.9 | 7.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.9 | 9.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.7 | 6.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.7 | 2.8 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.5 | 4.0 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 46.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.4 | 1.8 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.3 | 0.9 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.3 | 0.8 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.3 | 1.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 1.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 1.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 2.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.6 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 3.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.9 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 4.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.3 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.1 | 1.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 7.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.5 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 1.1 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 11.0 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 2.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 1.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 2.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 6.1 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 3.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 3.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 3.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 4.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.2 | 7.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.8 | 3.9 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.6 | 2.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.6 | 1.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.6 | 1.8 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.6 | 2.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.5 | 1.5 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.5 | 5.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.4 | 11.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 6.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.4 | 1.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 17.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.4 | 7.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 1.1 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.3 | 1.0 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.3 | 3.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 2.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 3.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.3 | 2.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 1.8 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 0.7 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 0.6 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.2 | 0.8 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 0.6 | GO:0035731 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.2 | 5.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 3.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 0.8 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 3.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 0.7 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 0.6 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.7 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 4.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.7 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 4.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.5 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 7.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 1.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 3.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 2.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 3.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.3 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.1 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 1.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 1.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 2.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 31.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 3.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 3.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 1.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 7.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 2.1 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 1.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 1.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 18.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 13.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 7.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 2.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 3.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 39.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 10.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 11.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 6.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 4.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 5.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 3.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |