Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Mafa
Z-value: 0.60
Transcription factors associated with Mafa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mafa
|
ENSMUSG00000047591.6 | Mafa |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafa | mm39_v1_chr15_-_75620060_75620087 | 0.17 | 1.4e-01 | Click! |
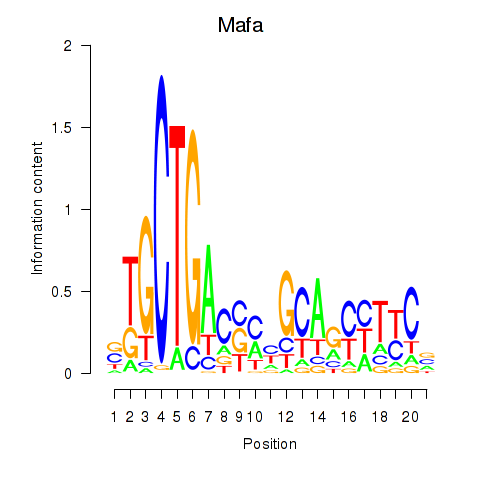
Activity profile of Mafa motif
Sorted Z-values of Mafa motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Mafa
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_135095344 | 6.69 |
ENSMUST00000027682.9
|
Gpr37l1
|
G protein-coupled receptor 37-like 1 |
| chr7_+_54485336 | 6.66 |
ENSMUST00000082373.8
|
Luzp2
|
leucine zipper protein 2 |
| chr8_-_70939964 | 6.58 |
ENSMUST00000045286.9
|
Tmem59l
|
transmembrane protein 59-like |
| chr1_-_169575203 | 6.12 |
ENSMUST00000027991.12
ENSMUST00000111357.2 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr6_-_35285045 | 4.50 |
ENSMUST00000031868.5
|
Slc13a4
|
solute carrier family 13 (sodium/sulfate symporters), member 4 |
| chr17_-_27732364 | 4.43 |
ENSMUST00000118161.3
|
Grm4
|
glutamate receptor, metabotropic 4 |
| chr9_-_53882530 | 4.37 |
ENSMUST00000048409.14
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr16_+_96268806 | 4.28 |
ENSMUST00000061739.9
|
Pcp4
|
Purkinje cell protein 4 |
| chr12_+_3415107 | 3.64 |
ENSMUST00000220210.2
|
Kif3c
|
kinesin family member 3C |
| chr4_+_102112189 | 3.64 |
ENSMUST00000106908.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_73605684 | 3.59 |
ENSMUST00000112024.10
ENSMUST00000180045.8 |
Chn1
|
chimerin 1 |
| chr4_+_102111936 | 3.13 |
ENSMUST00000106907.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr9_-_97915227 | 2.80 |
ENSMUST00000035027.13
|
Clstn2
|
calsyntenin 2 |
| chr11_-_66416621 | 2.66 |
ENSMUST00000123454.8
|
Shisa6
|
shisa family member 6 |
| chr13_+_97061182 | 2.57 |
ENSMUST00000171324.3
|
Gcnt4
|
glucosaminyl (N-acetyl) transferase 4, core 2 (beta-1,6-N-acetylglucosaminyltransferase) |
| chr6_+_85164420 | 2.55 |
ENSMUST00000045942.9
|
Emx1
|
empty spiracles homeobox 1 |
| chrX_+_150961960 | 2.49 |
ENSMUST00000096275.5
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr2_-_73605387 | 2.49 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr9_-_97915036 | 2.45 |
ENSMUST00000162295.2
|
Clstn2
|
calsyntenin 2 |
| chr11_+_104023657 | 2.33 |
ENSMUST00000093925.5
|
Crhr1
|
corticotropin releasing hormone receptor 1 |
| chr6_-_23839419 | 2.27 |
ENSMUST00000115358.9
ENSMUST00000163871.9 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chrX_-_20816841 | 2.18 |
ENSMUST00000009550.14
|
Elk1
|
ELK1, member of ETS oncogene family |
| chr5_+_111565129 | 2.14 |
ENSMUST00000094463.5
|
Mn1
|
meningioma 1 |
| chr11_-_107228382 | 1.96 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr11_-_76514334 | 1.92 |
ENSMUST00000238684.2
|
Abr
|
active BCR-related gene |
| chr11_-_100084072 | 1.86 |
ENSMUST00000059707.3
|
Krt9
|
keratin 9 |
| chr16_-_74208395 | 1.83 |
ENSMUST00000227347.2
|
Robo2
|
roundabout guidance receptor 2 |
| chr16_+_17149235 | 1.74 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr1_+_171041539 | 1.65 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr17_-_72910822 | 1.64 |
ENSMUST00000086639.6
|
Alk
|
anaplastic lymphoma kinase |
| chr14_-_24053994 | 1.56 |
ENSMUST00000225431.2
ENSMUST00000188210.8 ENSMUST00000224787.2 ENSMUST00000225315.2 ENSMUST00000225556.2 ENSMUST00000223727.2 ENSMUST00000223655.2 ENSMUST00000224077.2 ENSMUST00000224812.2 ENSMUST00000224285.2 ENSMUST00000225471.2 ENSMUST00000224232.2 ENSMUST00000223749.2 ENSMUST00000224025.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr8_-_85663976 | 1.53 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr16_-_74208180 | 1.45 |
ENSMUST00000117200.8
|
Robo2
|
roundabout guidance receptor 2 |
| chr8_+_94537910 | 1.45 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr12_-_75224099 | 1.44 |
ENSMUST00000042299.4
|
Kcnh5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| chr11_-_119119287 | 1.39 |
ENSMUST00000207655.2
ENSMUST00000036113.4 |
Tbc1d16
|
TBC1 domain family, member 16 |
| chr1_-_121255448 | 1.38 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr4_-_35845204 | 1.29 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr16_-_17394495 | 1.28 |
ENSMUST00000231645.2
ENSMUST00000232226.2 ENSMUST00000232336.2 ENSMUST00000232385.2 ENSMUST00000231615.2 ENSMUST00000231283.2 ENSMUST00000172164.10 |
Slc7a4
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 4 |
| chr12_+_3415143 | 1.27 |
ENSMUST00000020999.7
|
Kif3c
|
kinesin family member 3C |
| chr5_-_124018055 | 1.22 |
ENSMUST00000164267.2
|
Hcar1
|
hydrocarboxylic acid receptor 1 |
| chrX_-_101950075 | 1.21 |
ENSMUST00000120808.8
ENSMUST00000121197.8 |
Dmrtc1a
|
DMRT-like family C1a |
| chr14_-_24054273 | 1.21 |
ENSMUST00000188285.7
ENSMUST00000190044.7 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr4_+_155685830 | 1.17 |
ENSMUST00000105608.9
|
Slc35e2
|
solute carrier family 35, member E2 |
| chr4_+_155686311 | 1.16 |
ENSMUST00000118607.2
|
Slc35e2
|
solute carrier family 35, member E2 |
| chr4_+_43406435 | 1.11 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr8_-_106292616 | 1.02 |
ENSMUST00000013304.8
|
Atp6v0d1
|
ATPase, H+ transporting, lysosomal V0 subunit D1 |
| chr1_-_121255503 | 0.90 |
ENSMUST00000160688.2
|
Insig2
|
insulin induced gene 2 |
| chr1_-_121255400 | 0.89 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr5_-_24628514 | 0.71 |
ENSMUST00000030814.11
|
Cdk5
|
cyclin-dependent kinase 5 |
| chr7_+_17799863 | 0.70 |
ENSMUST00000108487.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr7_+_35148461 | 0.67 |
ENSMUST00000118969.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr14_+_79825131 | 0.63 |
ENSMUST00000054908.10
|
Sugt1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr5_-_24628483 | 0.61 |
ENSMUST00000198990.2
|
Cdk5
|
cyclin-dependent kinase 5 |
| chr4_+_117706390 | 0.60 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr1_-_121255753 | 0.58 |
ENSMUST00000003818.14
|
Insig2
|
insulin induced gene 2 |
| chr8_+_61446221 | 0.57 |
ENSMUST00000120689.8
ENSMUST00000034065.14 ENSMUST00000211256.2 ENSMUST00000211672.2 |
Nek1
|
NIMA (never in mitosis gene a)-related expressed kinase 1 |
| chr7_+_17799889 | 0.57 |
ENSMUST00000108483.2
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr6_-_57340712 | 0.56 |
ENSMUST00000228156.2
ENSMUST00000227186.2 ENSMUST00000228294.2 ENSMUST00000228342.2 |
Vmn1r17
|
vomeronasal 1 receptor 17 |
| chr6_-_69020489 | 0.53 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr16_+_32462927 | 0.51 |
ENSMUST00000124585.2
|
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr19_-_59158488 | 0.51 |
ENSMUST00000172821.4
|
Vax1
|
ventral anterior homeobox 1 |
| chr5_+_124767114 | 0.50 |
ENSMUST00000037865.13
|
Atp6v0a2
|
ATPase, H+ transporting, lysosomal V0 subunit A2 |
| chr19_+_27194757 | 0.49 |
ENSMUST00000047645.13
ENSMUST00000167487.8 |
Vldlr
|
very low density lipoprotein receptor |
| chr1_+_171041583 | 0.48 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chrX_+_103526380 | 0.47 |
ENSMUST00000087867.6
|
Uprt
|
uracil phosphoribosyltransferase |
| chr11_+_87486472 | 0.47 |
ENSMUST00000134216.3
|
Mtmr4
|
myotubularin related protein 4 |
| chr7_+_35148188 | 0.47 |
ENSMUST00000118383.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr7_+_35148579 | 0.45 |
ENSMUST00000032703.10
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr6_+_65358278 | 0.42 |
ENSMUST00000170608.8
|
Qrfprl
|
pyroglutamylated RFamide peptide receptor like |
| chr12_+_53294940 | 0.41 |
ENSMUST00000223358.3
|
Npas3
|
neuronal PAS domain protein 3 |
| chr7_+_17799849 | 0.39 |
ENSMUST00000032520.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chrX_-_72913410 | 0.38 |
ENSMUST00000066576.12
ENSMUST00000114430.8 |
L1cam
|
L1 cell adhesion molecule |
| chr19_+_27194411 | 0.37 |
ENSMUST00000164746.8
ENSMUST00000172302.8 |
Vldlr
|
very low density lipoprotein receptor |
| chr8_+_106099894 | 0.37 |
ENSMUST00000160650.2
|
Plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr9_+_21746785 | 0.37 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr9_+_108660989 | 0.35 |
ENSMUST00000192307.6
ENSMUST00000193560.6 ENSMUST00000194875.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr18_-_42712717 | 0.32 |
ENSMUST00000054738.5
|
Gpr151
|
G protein-coupled receptor 151 |
| chr7_-_12418918 | 0.29 |
ENSMUST00000167771.2
ENSMUST00000172743.9 ENSMUST00000233763.2 ENSMUST00000232880.2 |
Vmn2r55
|
vomeronasal 2, receptor 55 |
| chr3_-_153650068 | 0.28 |
ENSMUST00000150070.2
|
Acadm
|
acyl-Coenzyme A dehydrogenase, medium chain |
| chr13_+_55593116 | 0.25 |
ENSMUST00000001115.16
ENSMUST00000224995.2 ENSMUST00000225925.2 ENSMUST00000099482.5 ENSMUST00000224118.2 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr8_+_94863825 | 0.25 |
ENSMUST00000034207.8
|
Mt4
|
metallothionein 4 |
| chr17_-_47922374 | 0.19 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chr6_+_119213541 | 0.18 |
ENSMUST00000186622.3
|
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr4_-_141143313 | 0.16 |
ENSMUST00000006378.9
ENSMUST00000105788.2 |
Clcnkb
|
chloride channel, voltage-sensitive Kb |
| chr4_-_116321348 | 0.15 |
ENSMUST00000106486.8
ENSMUST00000106485.8 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr6_+_119213468 | 0.13 |
ENSMUST00000238905.2
ENSMUST00000037434.13 |
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr17_+_47922497 | 0.13 |
ENSMUST00000024778.3
|
Med20
|
mediator complex subunit 20 |
| chr6_-_69220672 | 0.10 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr3_-_153650269 | 0.06 |
ENSMUST00000072697.13
|
Acadm
|
acyl-Coenzyme A dehydrogenase, medium chain |
| chr19_-_36097233 | 0.06 |
ENSMUST00000025718.10
|
Ankrd1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr16_-_17394080 | 0.06 |
ENSMUST00000231552.2
|
Slc7a4
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 4 |
| chr9_-_105009023 | 0.01 |
ENSMUST00000035179.9
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr9_-_105008972 | 0.00 |
ENSMUST00000186925.2
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.6 | 2.3 | GO:0051866 | general adaptation syndrome(GO:0051866) regulation of corticosterone secretion(GO:2000852) |
| 0.5 | 1.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.5 | 6.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.5 | 2.6 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.4 | 4.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.4 | 1.6 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.3 | 2.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.3 | 1.7 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.3 | 6.7 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.3 | 2.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 1.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 1.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 | 4.3 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.2 | 0.6 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 3.7 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 4.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 0.6 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 2.6 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 2.5 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 7.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.9 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 2.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.5 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 6.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 2.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.5 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 6.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 4.9 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 4.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 1.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 1.4 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.4 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.2 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 6.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 4.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.3 | 1.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 4.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 6.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.5 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 3.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.9 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 2.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.2 | GO:0043679 | axon terminus(GO:0043679) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) corticotropin-releasing hormone binding(GO:0051424) |
| 0.6 | 4.4 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.5 | 2.6 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.4 | 1.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.3 | 3.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 0.9 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.2 | 1.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 1.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 1.3 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.2 | 1.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 2.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 2.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 6.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 6.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 6.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.3 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 4.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 6.7 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.5 | GO:0046961 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 7.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 3.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 8.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 1.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 8.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 6.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 3.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 2.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 8.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |