Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Mafb
Z-value: 1.44
Transcription factors associated with Mafb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mafb
|
ENSMUSG00000074622.5 | Mafb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafb | mm39_v1_chr2_-_160208977_160208993 | -0.01 | 9.4e-01 | Click! |
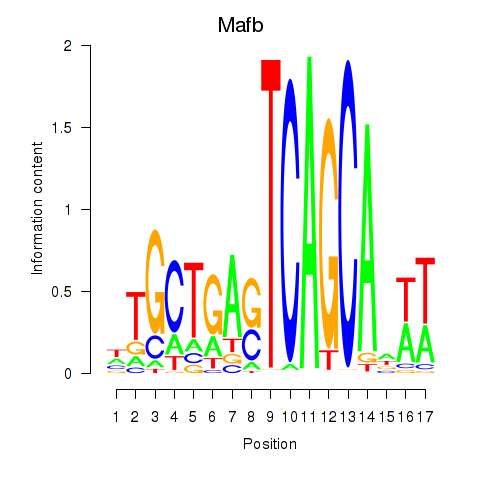
Activity profile of Mafb motif
Sorted Z-values of Mafb motif
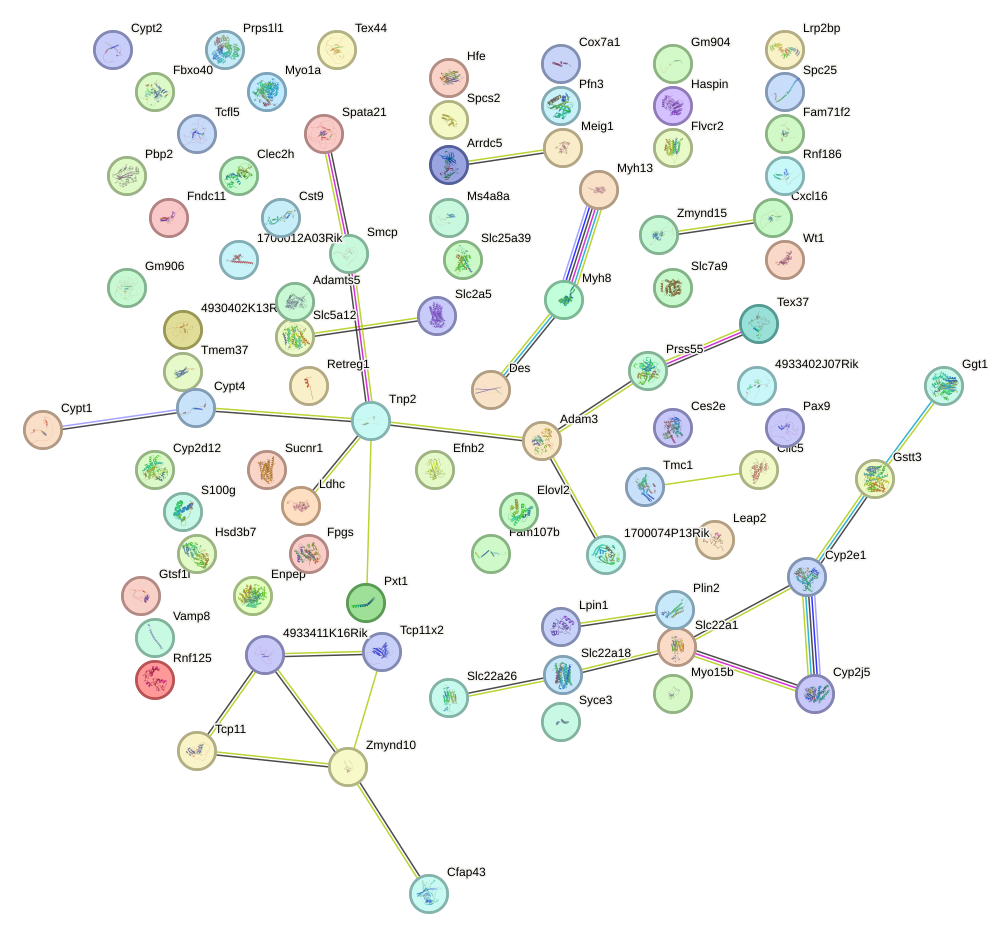
Network of associatons between targets according to the STRING database.
First level regulatory network of Mafb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_92496293 | 16.37 |
ENSMUST00000098888.7
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr17_-_28299569 | 13.02 |
ENSMUST00000129046.9
ENSMUST00000043925.16 |
Tcp11
|
t-complex protein 11 |
| chr4_-_96552349 | 11.74 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr7_+_46510677 | 10.20 |
ENSMUST00000211784.2
ENSMUST00000210585.2 ENSMUST00000148565.8 |
Ldhc
|
lactate dehydrogenase C |
| chr7_+_46510627 | 10.15 |
ENSMUST00000014545.11
|
Ldhc
|
lactate dehydrogenase C |
| chr17_-_15198955 | 9.87 |
ENSMUST00000231584.2
ENSMUST00000097399.6 ENSMUST00000232173.2 |
Dynlt2a3
|
dynein light chain Tctex-type 2A3 |
| chr17_-_15261701 | 9.54 |
ENSMUST00000097398.12
ENSMUST00000040746.8 ENSMUST00000097400.5 |
Dynlt2a1
Dynlt2a2
|
dynein light chain Tctex-type 2A1 dynein light chain Tctex-type 2A2 |
| chr7_+_46510831 | 9.21 |
ENSMUST00000126004.3
|
Ldhc
|
lactate dehydrogenase C |
| chr4_+_150203740 | 8.55 |
ENSMUST00000030826.4
|
Slc2a5
|
solute carrier family 2 (facilitated glucose transporter), member 5 |
| chr16_-_10606513 | 8.23 |
ENSMUST00000051297.9
|
Tnp2
|
transition protein 2 |
| chr6_+_128639342 | 7.71 |
ENSMUST00000032518.7
ENSMUST00000204416.2 |
Clec2h
|
C-type lectin domain family 2, member h |
| chr8_+_88290469 | 7.01 |
ENSMUST00000093342.6
|
4933402J07Rik
|
RIKEN cDNA 4933402J07 gene |
| chr11_+_115768323 | 6.95 |
ENSMUST00000222123.2
|
Myo15b
|
myosin XVB |
| chr2_+_148677060 | 6.91 |
ENSMUST00000028935.4
|
Cst9
|
cystatin 9 |
| chr19_+_42040681 | 6.91 |
ENSMUST00000164518.4
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene |
| chr1_+_75336965 | 6.70 |
ENSMUST00000027409.10
|
Des
|
desmin |
| chr1_-_120001752 | 6.63 |
ENSMUST00000056089.8
|
Tmem37
|
transmembrane protein 37 |
| chr11_-_73029070 | 6.46 |
ENSMUST00000052140.3
|
Haspin
|
histone H3 associated protein kinase |
| chr9_+_24536487 | 6.35 |
ENSMUST00000053956.8
|
Cypt4
|
cysteine-rich perinuclear theca 4 |
| chr2_-_180284468 | 6.23 |
ENSMUST00000037877.11
|
Tcfl5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr17_-_12894716 | 6.10 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr8_-_25215778 | 5.94 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr2_+_180861804 | 5.92 |
ENSMUST00000050026.7
ENSMUST00000108835.2 |
Fndc11
|
fibronectin type III domain containing 11 |
| chr13_-_55563028 | 5.92 |
ENSMUST00000054146.5
|
Pfn3
|
profilin 3 |
| chr12_+_35034747 | 5.84 |
ENSMUST00000134550.3
|
Prps1l1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr11_+_67167950 | 5.80 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr2_-_162929732 | 5.53 |
ENSMUST00000094653.6
|
Gtsf1l
|
gametocyte specific factor 1-like |
| chr2_+_104961228 | 5.53 |
ENSMUST00000111098.8
ENSMUST00000111099.2 |
Wt1
|
Wilms tumor 1 homolog |
| chr7_+_143028831 | 5.45 |
ENSMUST00000105917.3
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr8_-_25215856 | 5.41 |
ENSMUST00000033958.15
|
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr4_+_140815644 | 5.34 |
ENSMUST00000051907.3
|
Spata21
|
spermatogenesis associated 21 |
| chr16_-_36810810 | 5.27 |
ENSMUST00000075869.13
|
Fbxo40
|
F-box protein 40 |
| chrX_+_156482116 | 5.23 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr7_+_140343652 | 5.05 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr10_+_75409282 | 5.04 |
ENSMUST00000006508.10
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr8_+_105652867 | 4.86 |
ENSMUST00000034355.11
ENSMUST00000109410.4 |
Ces2e
|
carboxylesterase 2E |
| chr11_-_102298281 | 4.83 |
ENSMUST00000107098.8
ENSMUST00000018821.9 |
Slc25a39
|
solute carrier family 25, member 39 |
| chrX_-_161747552 | 4.79 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr13_-_41373638 | 4.73 |
ENSMUST00000117096.2
|
Elovl2
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
| chr2_-_60552980 | 4.62 |
ENSMUST00000028348.9
ENSMUST00000112517.8 |
Itgb6
|
integrin beta 6 |
| chr14_-_20502285 | 4.60 |
ENSMUST00000056073.14
ENSMUST00000022349.14 ENSMUST00000022348.15 |
Cfap70
|
cilia and flagella associated protein 70 |
| chr17_+_44499451 | 4.53 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr12_+_85793313 | 4.53 |
ENSMUST00000040461.4
|
Flvcr2
|
feline leukemia virus subgroup C cellular receptor 2 |
| chr3_-_51184895 | 4.46 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr19_-_7779943 | 4.46 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr3_-_129126362 | 4.40 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr11_+_70350252 | 4.38 |
ENSMUST00000108563.9
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr11_-_102298141 | 4.33 |
ENSMUST00000149777.8
ENSMUST00000154001.8 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr1_+_64729950 | 4.24 |
ENSMUST00000187170.7
|
Ccnyl1
|
cyclin Y-like 1 |
| chr6_-_70895899 | 4.24 |
ENSMUST00000063456.5
|
Tex37
|
testis expressed 37 |
| chr17_-_56607250 | 4.23 |
ENSMUST00000233911.2
|
Arrdc5
|
arrestin domain containing 5 |
| chr19_-_7780025 | 4.22 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chrX_+_16389108 | 4.21 |
ENSMUST00000115422.2
ENSMUST00000024026.3 |
Cypt1
|
cysteine-rich perinuclear theca 1 |
| chr6_+_32027216 | 4.21 |
ENSMUST00000031778.6
|
1700012A03Rik
|
RIKEN cDNA 1700012A03 gene |
| chr19_-_11058452 | 4.15 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr10_+_127541039 | 4.13 |
ENSMUST00000079590.7
|
Myo1a
|
myosin IA |
| chr13_-_23894697 | 4.12 |
ENSMUST00000091707.13
ENSMUST00000006787.8 |
Hfe
|
homeostatic iron regulator |
| chr7_+_35148188 | 4.11 |
ENSMUST00000118383.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr8_+_46463633 | 4.02 |
ENSMUST00000110381.9
|
Lrp2bp
|
Lrp2 binding protein |
| chr10_-_62198043 | 4.01 |
ENSMUST00000149534.2
|
Hk1
|
hexokinase 1 |
| chr3_+_40905216 | 4.01 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr6_-_40917431 | 3.96 |
ENSMUST00000122181.8
ENSMUST00000031935.10 |
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
| chr7_-_97387145 | 3.96 |
ENSMUST00000084986.8
|
Aqp11
|
aquaporin 11 |
| chr11_+_70350436 | 3.85 |
ENSMUST00000039093.10
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr11_-_102297590 | 3.83 |
ENSMUST00000155104.8
ENSMUST00000130436.8 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr9_+_107424488 | 3.83 |
ENSMUST00000010188.9
|
Zmynd10
|
zinc finger, MYND domain containing 10 |
| chr2_-_69036489 | 3.80 |
ENSMUST00000127243.8
ENSMUST00000149643.2 ENSMUST00000167875.9 ENSMUST00000005365.15 |
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr1_+_86354045 | 3.79 |
ENSMUST00000046004.6
|
Tex44
|
testis expressed 44 |
| chr13_+_24925192 | 3.76 |
ENSMUST00000136906.2
|
Armh2
|
armadillo-like helical domain containing 2 |
| chr15_-_89294434 | 3.74 |
ENSMUST00000109314.9
|
Syce3
|
synaptonemal complex central element protein 3 |
| chr13_+_50797264 | 3.72 |
ENSMUST00000099519.2
|
Gm904
|
predicted gene 904 |
| chr8_-_84738761 | 3.71 |
ENSMUST00000191523.2
ENSMUST00000190457.2 ENSMUST00000185457.2 |
Misp3
|
MISP family member 3 |
| chr6_-_135287372 | 3.71 |
ENSMUST00000050471.4
|
Pbp2
|
phosphatidylethanolamine binding protein 2 |
| chr12_+_56742413 | 3.71 |
ENSMUST00000001538.10
|
Pax9
|
paired box 9 |
| chr18_+_21094477 | 3.71 |
ENSMUST00000234316.2
|
Rnf125
|
ring finger protein 125 |
| chr10_-_75617245 | 3.70 |
ENSMUST00000001715.10
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr12_-_16696958 | 3.70 |
ENSMUST00000238839.2
|
Lpin1
|
lipin 1 |
| chr11_+_48728291 | 3.69 |
ENSMUST00000046903.6
|
Trim7
|
tripartite motif-containing 7 |
| chr2_+_110427643 | 3.69 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr7_-_97387429 | 3.66 |
ENSMUST00000206389.2
|
Aqp11
|
aquaporin 11 |
| chrX_+_156481906 | 3.63 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr2_-_3420056 | 3.55 |
ENSMUST00000115084.8
ENSMUST00000115083.8 |
Meig1
|
meiosis expressed gene 1 |
| chr9_+_44309727 | 3.51 |
ENSMUST00000213268.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr2_-_32584132 | 3.49 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr1_-_92830604 | 3.46 |
ENSMUST00000160548.8
ENSMUST00000112998.2 |
Ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr4_+_138694422 | 3.45 |
ENSMUST00000116094.5
ENSMUST00000239443.2 |
Rnf186
|
ring finger protein 186 |
| chr7_+_127399789 | 3.43 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_-_3420102 | 3.42 |
ENSMUST00000115082.10
|
Meig1
|
meiosis expressed gene 1 |
| chr7_+_29883611 | 3.38 |
ENSMUST00000208441.2
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr15_+_82439273 | 3.36 |
ENSMUST00000229103.2
ENSMUST00000068861.8 ENSMUST00000229904.2 |
Cyp2d12
|
cytochrome P450, family 2, subfamily d, polypeptide 12 |
| chr11_+_119119925 | 3.35 |
ENSMUST00000053440.8
|
Ccdc40
|
coiled-coil domain containing 40 |
| chr6_-_72367642 | 3.34 |
ENSMUST00000059983.10
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr2_+_104956850 | 3.31 |
ENSMUST00000143043.8
|
Wt1
|
Wilms tumor 1 homolog |
| chr14_-_64322849 | 3.31 |
ENSMUST00000089338.6
ENSMUST00000171503.8 |
Prss55
|
protease, serine 55 |
| chrX_+_104543374 | 3.30 |
ENSMUST00000041758.5
|
Cypt2
|
cysteine-rich perinuclear theca 2 |
| chr19_-_47825790 | 3.30 |
ENSMUST00000160247.3
|
Cfap43
|
cilia and flagella associated protein 43 |
| chr5_-_15734220 | 3.30 |
ENSMUST00000196384.2
|
Gm21190
|
predicted gene, 21190 |
| chr13_-_23894828 | 3.24 |
ENSMUST00000091706.14
|
Hfe
|
homeostatic iron regulator |
| chr4_-_86587728 | 3.24 |
ENSMUST00000149700.8
|
Plin2
|
perilipin 2 |
| chr11_-_53313950 | 3.22 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr15_+_25843225 | 3.21 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr6_+_29279582 | 3.20 |
ENSMUST00000167131.8
|
Fam71f2
|
family with sequence similarity 71, member F2 |
| chr19_-_20931566 | 3.18 |
ENSMUST00000039500.4
|
Tmc1
|
transmembrane channel-like gene family 1 |
| chrX_+_8970801 | 3.16 |
ENSMUST00000059967.2
|
4930402K13Rik
|
RIKEN cDNA 4930402K13 gene |
| chr3_+_59989282 | 3.15 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr13_-_50404344 | 3.14 |
ENSMUST00000099521.2
|
Gm906
|
predicted gene 906 |
| chr2_+_3714492 | 3.14 |
ENSMUST00000027965.11
|
Fam107b
|
family with sequence similarity 107, member B |
| chr14_-_54647647 | 3.12 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr17_-_29161229 | 3.12 |
ENSMUST00000051526.6
ENSMUST00000233413.2 |
Pxt1
|
peroxisomal, testis specific 1 |
| chr16_-_85698679 | 3.10 |
ENSMUST00000023611.7
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2) |
| chr11_+_75542328 | 3.07 |
ENSMUST00000069057.13
|
Myo1c
|
myosin IC |
| chr2_-_69036472 | 3.06 |
ENSMUST00000112320.8
|
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr5_-_14964913 | 3.04 |
ENSMUST00000035980.9
|
Gm9758
|
predicted gene 9758 |
| chr15_-_100579450 | 3.04 |
ENSMUST00000230740.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr5_-_139805661 | 3.02 |
ENSMUST00000147328.2
|
Tmem184a
|
transmembrane protein 184a |
| chr14_-_54648057 | 3.00 |
ENSMUST00000200545.2
ENSMUST00000227967.2 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr2_+_3714515 | 2.99 |
ENSMUST00000115053.9
|
Fam107b
|
family with sequence similarity 107, member B |
| chr5_-_15919234 | 2.97 |
ENSMUST00000095005.12
ENSMUST00000179506.2 |
Speer4c
|
spermatogenesis associated glutamate (E)-rich protein 4C |
| chr5_-_107873883 | 2.97 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr6_+_29471436 | 2.96 |
ENSMUST00000171317.2
|
Atp6v1fnb
|
Atp6v1f neighbor |
| chr11_-_69696428 | 2.96 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr15_-_5093222 | 2.94 |
ENSMUST00000110689.5
|
C7
|
complement component 7 |
| chr5_-_15681521 | 2.94 |
ENSMUST00000197995.2
|
Gm21149
|
predicted gene, 21149 |
| chr15_-_100579813 | 2.93 |
ENSMUST00000230572.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chrX_-_78715030 | 2.92 |
ENSMUST00000052283.7
|
Mageb16
|
MAGE family member B16 |
| chr1_-_121255503 | 2.91 |
ENSMUST00000160688.2
|
Insig2
|
insulin induced gene 2 |
| chr1_+_167177545 | 2.90 |
ENSMUST00000028004.11
|
Aldh9a1
|
aldehyde dehydrogenase 9, subfamily A1 |
| chr12_+_65272495 | 2.83 |
ENSMUST00000221980.2
|
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr19_-_20704896 | 2.81 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr11_-_94398162 | 2.81 |
ENSMUST00000040692.9
|
Mycbpap
|
MYCBP associated protein |
| chr1_-_121255753 | 2.79 |
ENSMUST00000003818.14
|
Insig2
|
insulin induced gene 2 |
| chr5_+_15782286 | 2.77 |
ENSMUST00000188807.4
|
Gm21083
|
predicted gene, 21083 |
| chr15_+_82338247 | 2.74 |
ENSMUST00000230000.2
|
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr1_-_161807205 | 2.70 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr5_-_121756928 | 2.67 |
ENSMUST00000041252.13
ENSMUST00000111776.6 |
Acad12
|
acyl-Coenzyme A dehydrogenase family, member 12 |
| chr2_-_27136826 | 2.63 |
ENSMUST00000149733.8
|
Sardh
|
sarcosine dehydrogenase |
| chr7_+_140427711 | 2.62 |
ENSMUST00000026555.12
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr9_+_45314436 | 2.61 |
ENSMUST00000041005.6
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr15_+_31224616 | 2.60 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr7_+_40682143 | 2.59 |
ENSMUST00000164422.2
|
Gm4884
|
predicted gene 4884 |
| chr8_+_117822593 | 2.58 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr5_-_15028949 | 2.56 |
ENSMUST00000096953.5
|
Gm10354
|
predicted gene 10354 |
| chr15_+_25933632 | 2.54 |
ENSMUST00000228327.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr8_-_123302187 | 2.53 |
ENSMUST00000213062.2
|
Aprt
|
adenine phosphoribosyl transferase |
| chr2_+_164131181 | 2.51 |
ENSMUST00000017147.8
ENSMUST00000109370.2 |
Svs3a
|
seminal vesicle secretory protein 3A |
| chr5_-_15083012 | 2.48 |
ENSMUST00000167908.2
|
Gm17019
|
predicted gene 17019 |
| chr12_-_16660960 | 2.48 |
ENSMUST00000239165.2
ENSMUST00000111067.10 |
Lpin1
|
lipin 1 |
| chr11_+_75542902 | 2.46 |
ENSMUST00000102504.10
|
Myo1c
|
myosin IC |
| chr15_-_59888446 | 2.46 |
ENSMUST00000096421.4
|
4933412E24Rik
|
RIKEN cDNA 4933412E24 gene |
| chr5_-_14988443 | 2.44 |
ENSMUST00000159973.3
|
Speer4e
|
spermatogenesis associated glutamate (E)-rich protein 4e |
| chr11_-_109364424 | 2.42 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr2_-_127673738 | 2.41 |
ENSMUST00000028858.8
|
Bub1
|
BUB1, mitotic checkpoint serine/threonine kinase |
| chr6_+_40763875 | 2.40 |
ENSMUST00000195870.3
|
Mgam2-ps
|
maltase-glucoamylase 2, pseudogene |
| chr14_+_22069709 | 2.40 |
ENSMUST00000075639.11
|
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr5_-_26193648 | 2.40 |
ENSMUST00000191203.7
|
Gm21698
|
predicted gene, 21698 |
| chr5_+_20112771 | 2.39 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_+_17578178 | 2.38 |
ENSMUST00000166086.3
|
Speer4f2
|
spermatogenesis associated glutamate (E)-rich protein 4f2 |
| chr11_-_116197994 | 2.37 |
ENSMUST00000124281.2
|
Exoc7
|
exocyst complex component 7 |
| chr1_+_64729603 | 2.37 |
ENSMUST00000114077.8
|
Ccnyl1
|
cyclin Y-like 1 |
| chr12_+_65272287 | 2.37 |
ENSMUST00000046331.5
ENSMUST00000221658.2 |
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr10_-_129738595 | 2.36 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr14_-_34032311 | 2.35 |
ENSMUST00000111917.3
ENSMUST00000228704.2 |
Shld2
|
shieldin complex subunit 2 |
| chr14_-_34310438 | 2.34 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr10_+_70040483 | 2.33 |
ENSMUST00000020090.8
|
Mrln
|
myoregulin |
| chr7_+_140427729 | 2.33 |
ENSMUST00000106049.2
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr5_+_17681096 | 2.32 |
ENSMUST00000076099.4
|
Speer4f1
|
spermatogenesis associated glutamate (E)-rich protein 4F1 |
| chr7_-_39062584 | 2.32 |
ENSMUST00000108017.2
|
Gm5114
|
predicted gene 5114 |
| chr1_+_54369151 | 2.28 |
ENSMUST00000163072.8
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr14_-_66361931 | 2.26 |
ENSMUST00000070515.2
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr1_-_121255400 | 2.25 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr5_-_15862057 | 2.24 |
ENSMUST00000092696.5
|
4930572O03Rik
|
RIKEN cDNA 4930572O03 gene |
| chr7_+_127399776 | 2.23 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr7_-_24423715 | 2.21 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr11_-_83412952 | 2.20 |
ENSMUST00000019069.4
|
Heatr9
|
HEAT repeat containing 9 |
| chr9_-_107556823 | 2.19 |
ENSMUST00000010205.9
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chr13_+_4283729 | 2.18 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr17_+_34617784 | 2.17 |
ENSMUST00000114175.8
ENSMUST00000139063.8 ENSMUST00000078615.12 |
BC051142
|
cDNA sequence BC051142 |
| chr7_-_30443106 | 2.17 |
ENSMUST00000182634.8
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr11_-_121009503 | 2.16 |
ENSMUST00000039146.4
|
Tex19.2
|
testis expressed gene 19.2 |
| chr15_-_75801575 | 2.16 |
ENSMUST00000229951.2
ENSMUST00000023231.7 ENSMUST00000230736.2 |
Gfus
|
GDP-L-fucose synthase |
| chr15_-_79212323 | 2.15 |
ENSMUST00000166977.9
|
Pla2g6
|
phospholipase A2, group VI |
| chr15_-_76193955 | 2.14 |
ENSMUST00000210024.2
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr11_+_106265660 | 2.12 |
ENSMUST00000188561.7
ENSMUST00000190795.7 ENSMUST00000185986.7 ENSMUST00000190268.2 |
Prr29
|
proline rich 29 |
| chr3_+_108444837 | 2.11 |
ENSMUST00000029485.6
|
1700013F07Rik
|
RIKEN cDNA 1700013F07 gene |
| chr11_-_102815910 | 2.10 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr19_-_44534274 | 2.07 |
ENSMUST00000111985.2
ENSMUST00000063632.14 |
Sec31b
|
Sec31 homolog B (S. cerevisiae) |
| chr5_-_27706360 | 2.04 |
ENSMUST00000155721.2
ENSMUST00000053257.10 |
Speer4b
|
spermatogenesis associated glutamate (E)-rich protein 4B |
| chr9_-_103165489 | 2.04 |
ENSMUST00000035163.10
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr11_+_82006001 | 2.02 |
ENSMUST00000009329.3
|
Ccl8
|
chemokine (C-C motif) ligand 8 |
| chrX_+_122925694 | 2.02 |
ENSMUST00000238503.2
|
Gm5945
|
predicted gene 5945 |
| chr15_-_56557920 | 2.02 |
ENSMUST00000050544.8
|
Has2
|
hyaluronan synthase 2 |
| chr3_+_14545751 | 1.98 |
ENSMUST00000037321.8
ENSMUST00000120484.8 ENSMUST00000120801.2 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr12_+_84085136 | 1.96 |
ENSMUST00000021652.5
|
Acot4
|
acyl-CoA thioesterase 4 |
| chr1_-_121255448 | 1.95 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr12_+_104082229 | 1.95 |
ENSMUST00000021496.8
|
Serpina3a
|
serine (or cysteine) peptidase inhibitor, clade A, member 3A |
| chr11_+_106265645 | 1.94 |
ENSMUST00000106816.8
|
Prr29
|
proline rich 29 |
| chrX_+_122256461 | 1.94 |
ENSMUST00000238410.2
|
Gm2411
|
predicted gene 2411 |
| chr15_-_75801630 | 1.94 |
ENSMUST00000229289.2
ENSMUST00000229641.2 |
Gfus
|
GDP-L-fucose synthase |
| chr2_-_152775122 | 1.93 |
ENSMUST00000099200.3
|
Foxs1
|
forkhead box S1 |
| chr4_+_98284054 | 1.93 |
ENSMUST00000107033.8
ENSMUST00000107034.8 |
Patj
|
PATJ, crumbs cell polarity complex component |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 29.6 | GO:0019516 | lactate oxidation(GO:0019516) |
| 3.9 | 11.7 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 3.3 | 13.0 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 3.1 | 24.6 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 2.9 | 8.8 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 2.5 | 7.4 | GO:0002590 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 2.2 | 6.5 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 2.1 | 8.6 | GO:0015755 | fructose transport(GO:0015755) |
| 1.9 | 7.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 1.2 | 3.5 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 1.1 | 4.2 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 1.0 | 4.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 1.0 | 6.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.9 | 2.6 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.9 | 6.0 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.8 | 3.3 | GO:1902276 | autophagosome docking(GO:0016240) pancreatic amylase secretion(GO:0036395) zymogen granule exocytosis(GO:0070625) positive regulation of pancreatic juice secretion(GO:0090187) regulation of pancreatic amylase secretion(GO:1902276) |
| 0.8 | 6.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.8 | 2.3 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.7 | 3.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.7 | 5.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.7 | 2.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.7 | 4.0 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.7 | 2.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.6 | 5.7 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.6 | 2.5 | GO:0006168 | adenine salvage(GO:0006168) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.6 | 5.0 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.6 | 4.4 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.6 | 2.5 | GO:0045297 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.6 | 9.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.6 | 6.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.6 | 2.4 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.6 | 9.9 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.5 | 3.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.5 | 1.6 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.5 | 1.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.5 | 6.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.5 | 4.6 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.5 | 3.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.5 | 8.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.5 | 7.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.5 | 2.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.4 | 1.7 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.4 | 4.7 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 3.5 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.4 | 1.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 | 1.9 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.4 | 2.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.4 | 13.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.3 | 1.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 3.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 0.9 | GO:0036034 | mediator complex assembly(GO:0036034) DNA replication preinitiation complex assembly(GO:0071163) response to sorbitol(GO:0072708) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.3 | 5.8 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.3 | 1.3 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.3 | 1.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 3.0 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.2 | 0.7 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.2 | 11.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 1.7 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 4.5 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.2 | 1.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 1.0 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.2 | 2.4 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 2.4 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 0.4 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.2 | 1.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 0.6 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.2 | 0.6 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 1.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 2.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 5.0 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.2 | 1.6 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 | 2.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.2 | 0.2 | GO:0050942 | positive regulation of developmental pigmentation(GO:0048087) regulation of pigment cell differentiation(GO:0050932) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.2 | 1.4 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 0.9 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 1.1 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.2 | 4.6 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 2.8 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 6.9 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.1 | 0.4 | GO:1904156 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.1 | 1.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.9 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.6 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 1.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 1.6 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 2.7 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 2.6 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 2.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 4.1 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 1.6 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.5 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 1.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 4.5 | GO:0015893 | drug transport(GO:0015893) |
| 0.1 | 1.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 9.0 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 0.7 | GO:0051608 | histamine transport(GO:0051608) |
| 0.1 | 3.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 3.7 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 6.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.8 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 0.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 4.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 6.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 2.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 1.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.4 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.5 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.9 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 2.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 5.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.4 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 36.0 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 0.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.2 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.0 | 0.7 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.7 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 1.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 1.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.5 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 1.0 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 3.2 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 1.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.8 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 3.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 1.9 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.9 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 1.1 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 1.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 4.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 4.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 2.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.7 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.6 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.7 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 6.9 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 1.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.4 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.9 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 6.5 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.5 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 1.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 2.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.5 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 1.3 | GO:0042303 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.1 | 6.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.1 | 8.9 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.1 | 5.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.9 | 4.6 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.8 | 7.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.8 | 2.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.6 | 6.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.6 | 14.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.4 | 13.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.3 | 3.7 | GO:0000801 | central element(GO:0000801) |
| 0.3 | 2.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 4.4 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.3 | 4.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 2.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 3.7 | GO:0005818 | aster(GO:0005818) |
| 0.3 | 0.8 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 1.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 2.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 5.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 3.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 1.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 6.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 1.3 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 36.8 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 3.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 2.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 6.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 22.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 5.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 4.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 9.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 23.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.4 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 3.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 3.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 2.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 5.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.7 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 1.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.0 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 4.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.5 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 2.7 | 29.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 2.0 | 6.1 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.6 | 6.6 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 1.6 | 6.5 | GO:0072354 | histone kinase activity (H3-T3 specific)(GO:0072354) |
| 1.3 | 8.8 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 1.2 | 3.5 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 1.2 | 3.5 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 1.0 | 4.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.9 | 2.6 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.8 | 2.5 | GO:0002055 | adenine binding(GO:0002055) adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.8 | 4.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.7 | 2.9 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.6 | 1.9 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.6 | 4.8 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.6 | 7.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.6 | 2.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.6 | 9.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 20.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.5 | 1.5 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.5 | 2.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.5 | 2.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.4 | 1.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.4 | 2.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 1.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 7.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 5.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 3.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.4 | 4.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 4.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 0.4 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.4 | 7.7 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.3 | 2.4 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.3 | 1.3 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.3 | 3.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 2.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 6.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 1.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.3 | 3.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 3.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 2.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 7.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 1.7 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 6.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.7 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.2 | 4.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 4.5 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.2 | 1.6 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 2.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 3.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 1.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 2.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 2.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 5.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 3.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 6.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 2.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 2.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 3.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.0 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 2.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 1.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 1.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.6 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 7.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 5.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 8.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 10.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.0 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 2.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 4.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 3.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 3.0 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 2.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.2 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 3.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 5.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 2.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 9.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 8.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 5.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 5.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 14.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 3.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.8 | 12.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.6 | 9.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 6.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 1.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.2 | 5.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 9.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 13.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 4.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 9.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 4.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 6.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 5.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 3.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 9.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.9 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 2.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 3.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 4.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.2 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 3.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |