Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
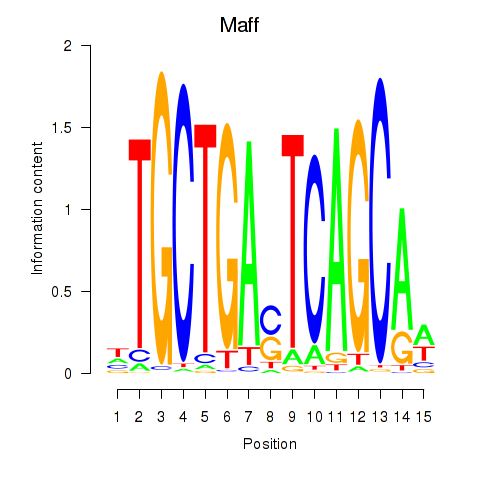
Results for Maff
Z-value: 0.74
Transcription factors associated with Maff
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Maff
|
ENSMUSG00000042622.15 | Maff |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Maff | mm39_v1_chr15_+_79231720_79231763 | -0.33 | 5.1e-03 | Click! |
Activity profile of Maff motif
Sorted Z-values of Maff motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Maff
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_119025306 | 8.19 |
ENSMUST00000047761.13
ENSMUST00000071546.14 |
Cldn10
|
claudin 10 |
| chr16_+_96295011 | 8.09 |
ENSMUST00000233816.2
|
Pcp4
|
Purkinje cell protein 4 |
| chr7_-_74958121 | 7.44 |
ENSMUST00000085164.7
|
Sv2b
|
synaptic vesicle glycoprotein 2 b |
| chr13_-_55677109 | 4.97 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3 |
| chr3_-_59170245 | 4.31 |
ENSMUST00000050360.14
ENSMUST00000199609.2 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr15_-_33687986 | 4.25 |
ENSMUST00000042021.5
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr7_-_64575308 | 4.02 |
ENSMUST00000149851.8
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr6_+_120750510 | 3.93 |
ENSMUST00000112682.4
|
Slc25a18
|
solute carrier family 25 (mitochondrial carrier), member 18 |
| chr3_-_80710097 | 3.83 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr18_+_13107535 | 3.73 |
ENSMUST00000234035.2
ENSMUST00000235053.2 |
Impact
|
impact, RWD domain protein |
| chr10_-_88440869 | 3.62 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr2_-_130239434 | 3.57 |
ENSMUST00000028897.8
|
Cpxm1
|
carboxypeptidase X 1 (M14 family) |
| chr7_+_16678568 | 3.51 |
ENSMUST00000094807.6
|
Pnmal2
|
PNMA-like 2 |
| chrX_+_92718695 | 3.24 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr5_+_140491305 | 3.16 |
ENSMUST00000043050.9
ENSMUST00000124142.2 |
Chst12
|
carbohydrate sulfotransferase 12 |
| chr7_-_108769719 | 3.02 |
ENSMUST00000208136.2
ENSMUST00000036992.9 |
Lmo1
|
LIM domain only 1 |
| chr5_-_67973195 | 2.99 |
ENSMUST00000141443.2
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr18_+_69654572 | 2.92 |
ENSMUST00000200862.4
|
Tcf4
|
transcription factor 4 |
| chr11_-_120520954 | 2.91 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr5_+_107112186 | 2.90 |
ENSMUST00000117196.9
ENSMUST00000031221.12 ENSMUST00000076467.13 |
Cdc7
|
cell division cycle 7 (S. cerevisiae) |
| chr8_+_124138163 | 2.88 |
ENSMUST00000071134.4
ENSMUST00000212743.2 |
Tubb3
|
tubulin, beta 3 class III |
| chr18_+_69654900 | 2.88 |
ENSMUST00000202057.4
|
Tcf4
|
transcription factor 4 |
| chr18_+_69654231 | 2.86 |
ENSMUST00000202350.4
ENSMUST00000202477.4 |
Tcf4
|
transcription factor 4 |
| chr6_+_4003904 | 2.83 |
ENSMUST00000031670.10
|
Gng11
|
guanine nucleotide binding protein (G protein), gamma 11 |
| chr3_-_108062172 | 2.78 |
ENSMUST00000062028.8
|
Gpr61
|
G protein-coupled receptor 61 |
| chr1_+_75362187 | 2.72 |
ENSMUST00000137868.8
|
Speg
|
SPEG complex locus |
| chr2_+_121697566 | 2.67 |
ENSMUST00000089912.12
ENSMUST00000089915.10 |
Golm2
|
golgi membrane protein 2 |
| chr10_-_88440996 | 2.55 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr2_+_121697398 | 2.51 |
ENSMUST00000110586.10
ENSMUST00000078752.10 |
Golm2
|
golgi membrane protein 2 |
| chr2_-_84717036 | 2.46 |
ENSMUST00000054514.6
ENSMUST00000151799.8 |
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr3_+_109481223 | 2.22 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr17_-_25789652 | 2.20 |
ENSMUST00000025003.10
ENSMUST00000173447.2 |
Sox8
|
SRY (sex determining region Y)-box 8 |
| chr8_+_26022141 | 2.08 |
ENSMUST00000210846.2
ENSMUST00000167764.2 |
Fgfr1
|
fibroblast growth factor receptor 1 |
| chr1_+_75358758 | 2.01 |
ENSMUST00000148515.8
ENSMUST00000113590.8 |
Speg
|
SPEG complex locus |
| chr14_+_15369152 | 1.83 |
ENSMUST00000167923.8
|
Gm3696
|
predicted gene 3696 |
| chr11_+_81992662 | 1.82 |
ENSMUST00000000194.4
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr4_+_102278715 | 1.73 |
ENSMUST00000106904.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr16_+_72460029 | 1.66 |
ENSMUST00000023600.8
|
Robo1
|
roundabout guidance receptor 1 |
| chr18_+_37606591 | 1.63 |
ENSMUST00000050034.3
|
Pcdhb15
|
protocadherin beta 15 |
| chr17_+_71511642 | 1.62 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr9_+_45749869 | 1.61 |
ENSMUST00000078111.11
ENSMUST00000034591.11 |
Bace1
|
beta-site APP cleaving enzyme 1 |
| chr11_-_120521382 | 1.59 |
ENSMUST00000106181.8
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr2_-_20948230 | 1.59 |
ENSMUST00000140230.2
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr4_+_139079898 | 1.52 |
ENSMUST00000179784.9
ENSMUST00000082262.8 ENSMUST00000042096.15 ENSMUST00000147999.8 |
Emc1
Ubr4
|
ER membrane protein complex subunit 1 ubiquitin protein ligase E3 component n-recognin 4 |
| chr19_+_53186430 | 1.50 |
ENSMUST00000237099.2
|
Add3
|
adducin 3 (gamma) |
| chr8_+_72050292 | 1.45 |
ENSMUST00000143662.8
|
Niban3
|
niban apoptosis regulator 3 |
| chr18_+_69654992 | 1.40 |
ENSMUST00000201627.4
|
Tcf4
|
transcription factor 4 |
| chr3_+_36205166 | 1.39 |
ENSMUST00000197653.5
ENSMUST00000205077.2 |
Zfp267
|
zinc finger protein 267 |
| chr7_+_30014235 | 1.34 |
ENSMUST00000054594.15
ENSMUST00000177078.8 ENSMUST00000176504.8 ENSMUST00000176304.8 |
Syne4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr13_-_54897660 | 1.24 |
ENSMUST00000135343.2
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chr11_-_119119287 | 1.22 |
ENSMUST00000207655.2
ENSMUST00000036113.4 |
Tbc1d16
|
TBC1 domain family, member 16 |
| chr1_+_135945705 | 1.19 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr4_-_136613498 | 1.16 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr13_-_54897425 | 1.15 |
ENSMUST00000099506.2
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chr11_+_83599841 | 1.15 |
ENSMUST00000001009.14
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr2_+_85606930 | 1.13 |
ENSMUST00000076250.2
|
Olfr1014
|
olfactory receptor 1014 |
| chr1_-_54234193 | 1.12 |
ENSMUST00000087659.11
ENSMUST00000097741.3 |
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr7_-_7302468 | 1.12 |
ENSMUST00000000619.8
|
Clcn4
|
chloride channel, voltage-sensitive 4 |
| chr8_-_12722099 | 1.02 |
ENSMUST00000000776.15
|
Tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr18_-_39652468 | 0.95 |
ENSMUST00000237944.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr17_+_28059129 | 0.93 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr17_+_28059099 | 0.88 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr7_+_44117444 | 0.86 |
ENSMUST00000206887.2
ENSMUST00000117324.8 ENSMUST00000120852.8 ENSMUST00000134398.3 ENSMUST00000118628.8 |
Josd2
|
Josephin domain containing 2 |
| chr7_-_7301760 | 0.81 |
ENSMUST00000210061.2
|
Clcn4
|
chloride channel, voltage-sensitive 4 |
| chr4_+_156214969 | 0.80 |
ENSMUST00000209248.2
|
Rnf223
|
ring finger 223 |
| chr3_+_28859585 | 0.75 |
ENSMUST00000043867.11
ENSMUST00000194649.2 |
Rpl22l1
|
ribosomal protein L22 like 1 |
| chr7_-_47818767 | 0.74 |
ENSMUST00000094389.4
|
Mrgprb5
|
MAS-related GPR, member B5 |
| chr11_+_76836330 | 0.73 |
ENSMUST00000021197.10
|
Blmh
|
bleomycin hydrolase |
| chr4_-_139079842 | 0.72 |
ENSMUST00000102503.10
|
Mrto4
|
mRNA turnover 4, ribosome maturation factor |
| chr2_-_152775122 | 0.71 |
ENSMUST00000099200.3
|
Foxs1
|
forkhead box S1 |
| chr9_+_65368207 | 0.71 |
ENSMUST00000034955.8
ENSMUST00000213957.2 |
Spg21
|
SPG21, maspardin |
| chr7_-_4687916 | 0.71 |
ENSMUST00000206306.2
ENSMUST00000205952.2 ENSMUST00000079970.6 |
Hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr19_-_57185988 | 0.70 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr5_-_102217770 | 0.64 |
ENSMUST00000053177.14
ENSMUST00000174698.2 |
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr8_-_84420633 | 0.63 |
ENSMUST00000144258.8
|
Pkn1
|
protein kinase N1 |
| chr14_-_20844074 | 0.63 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr1_+_72346572 | 0.63 |
ENSMUST00000027379.10
|
Xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 |
| chr7_+_101032021 | 0.61 |
ENSMUST00000141083.9
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr14_-_36857083 | 0.60 |
ENSMUST00000042564.17
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr17_+_38485977 | 0.57 |
ENSMUST00000074883.2
|
Olfr134
|
olfactory receptor 134 |
| chr17_+_35960600 | 0.57 |
ENSMUST00000171166.3
|
Sfta2
|
surfactant associated 2 |
| chr12_-_101943134 | 0.52 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr7_+_44117511 | 0.51 |
ENSMUST00000121922.3
ENSMUST00000208117.2 |
Josd2
|
Josephin domain containing 2 |
| chr4_-_139079609 | 0.50 |
ENSMUST00000030513.13
ENSMUST00000155257.8 |
Mrto4
|
mRNA turnover 4, ribosome maturation factor |
| chr1_+_127234441 | 0.49 |
ENSMUST00000171405.2
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr1_+_135945798 | 0.48 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr14_-_36857202 | 0.48 |
ENSMUST00000165649.4
ENSMUST00000224769.2 |
Ghitm
|
growth hormone inducible transmembrane protein |
| chr17_+_28059036 | 0.43 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr5_-_35683035 | 0.38 |
ENSMUST00000038676.7
|
Cpz
|
carboxypeptidase Z |
| chr6_+_40941688 | 0.37 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr14_+_20979466 | 0.37 |
ENSMUST00000022369.9
|
Vcl
|
vinculin |
| chr14_+_30853010 | 0.34 |
ENSMUST00000227096.2
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr10_+_62756426 | 0.33 |
ENSMUST00000144459.2
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr11_+_76836545 | 0.32 |
ENSMUST00000125145.8
|
Blmh
|
bleomycin hydrolase |
| chr5_+_8106527 | 0.31 |
ENSMUST00000148633.4
|
Sri
|
sorcin |
| chr14_+_55170152 | 0.30 |
ENSMUST00000037863.6
|
Il25
|
interleukin 25 |
| chr7_+_30193047 | 0.29 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr10_+_62756409 | 0.29 |
ENSMUST00000044977.10
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr7_+_138794577 | 0.28 |
ENSMUST00000135509.2
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr9_-_111519382 | 0.28 |
ENSMUST00000035083.8
|
Stac
|
src homology three (SH3) and cysteine rich domain |
| chrX_+_99568437 | 0.27 |
ENSMUST00000037541.9
|
Dgat2l6
|
diacylglycerol O-acyltransferase 2-like 6 |
| chr11_-_69471056 | 0.26 |
ENSMUST00000132548.2
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr5_-_24963006 | 0.21 |
ENSMUST00000047119.5
|
Crygn
|
crystallin, gamma N |
| chr13_+_22563988 | 0.21 |
ENSMUST00000227685.2
ENSMUST00000227689.2 ENSMUST00000227846.2 |
Vmn1r199
|
vomeronasal 1 receptor 199 |
| chr7_+_44117404 | 0.20 |
ENSMUST00000035844.11
|
Josd2
|
Josephin domain containing 2 |
| chr7_+_44117475 | 0.19 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chr9_-_78220729 | 0.17 |
ENSMUST00000085308.5
|
Omt2a
|
oocyte maturation, alpha |
| chr3_-_106126794 | 0.16 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr6_-_40976413 | 0.15 |
ENSMUST00000166306.3
|
Gm2663
|
predicted gene 2663 |
| chr13_+_67080864 | 0.15 |
ENSMUST00000021990.4
|
Ptdss1
|
phosphatidylserine synthase 1 |
| chr6_-_70036183 | 0.12 |
ENSMUST00000197429.5
ENSMUST00000103376.3 |
Igkv7-33
|
immunoglobulin kappa chain variable 7-33 |
| chr7_-_100613579 | 0.10 |
ENSMUST00000060174.6
|
P2ry6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr7_-_12771554 | 0.10 |
ENSMUST00000125964.8
|
Ube2m
|
ubiquitin-conjugating enzyme E2M |
| chr11_+_74721733 | 0.07 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein |
| chr6_+_83771953 | 0.06 |
ENSMUST00000037376.14
|
Nagk
|
N-acetylglucosamine kinase |
| chr18_-_38471962 | 0.03 |
ENSMUST00000139885.2
ENSMUST00000235590.2 ENSMUST00000237487.2 ENSMUST00000063814.15 |
Gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr3_+_108847375 | 0.03 |
ENSMUST00000197427.5
ENSMUST00000059946.11 ENSMUST00000098680.9 |
Henmt1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1901561 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 1.1 | 4.3 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.1 | 3.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.7 | 3.7 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.7 | 2.2 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.5 | 2.9 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.4 | 8.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.4 | 3.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.4 | 10.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 1.8 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.4 | 6.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 3.0 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 | 1.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 1.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.3 | 1.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 2.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 0.6 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.9 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 1.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 3.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 4.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.0 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.6 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 2.5 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.1 | 0.6 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 2.2 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 4.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.3 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.3 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 3.2 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 4.7 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 1.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 1.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 7.4 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 3.6 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.7 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 1.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 3.8 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 1.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 6.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 0.6 | GO:0097635 | Atg12-Atg5-Atg16 complex(GO:0034274) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.2 | 3.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 2.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.6 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 1.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.3 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.1 | 10.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 7.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 5.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 3.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 5.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.8 | 3.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.6 | 3.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.6 | 10.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.6 | 2.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.5 | 1.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 4.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 6.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 3.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 2.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.9 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 0.6 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 4.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 1.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 3.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.6 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 5.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 2.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 1.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 3.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 10.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 2.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 11.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 5.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 6.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 10.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 3.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 5.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.8 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 2.9 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 3.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |