Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Mef2b
Z-value: 1.44
Transcription factors associated with Mef2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2b
|
ENSMUSG00000079033.11 | Mef2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2b | mm39_v1_chr8_+_70592337_70592384 | 0.16 | 1.7e-01 | Click! |
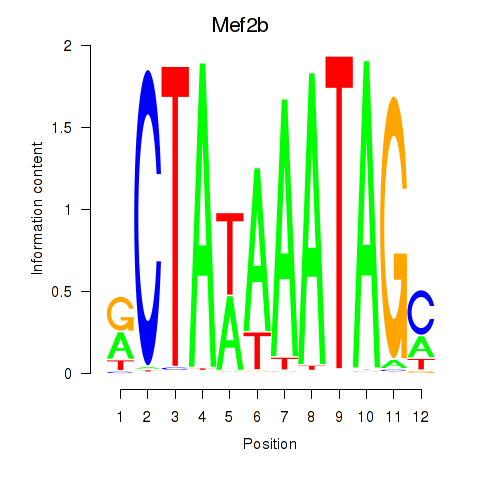
Activity profile of Mef2b motif
Sorted Z-values of Mef2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_61096660 | 10.79 |
ENSMUST00000039904.7
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr18_+_61096597 | 10.50 |
ENSMUST00000115295.9
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr7_-_127805518 | 9.94 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr4_+_46039202 | 9.45 |
ENSMUST00000156200.8
|
Tmod1
|
tropomodulin 1 |
| chr11_-_79394904 | 9.08 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr4_-_134099840 | 8.76 |
ENSMUST00000030643.3
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr8_-_46664321 | 8.43 |
ENSMUST00000034049.5
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr6_-_71239216 | 8.30 |
ENSMUST00000129630.3
ENSMUST00000114186.9 ENSMUST00000074301.10 |
Smyd1
|
SET and MYND domain containing 1 |
| chr9_+_121606750 | 8.21 |
ENSMUST00000098272.4
|
Klhl40
|
kelch-like 40 |
| chr15_-_98705791 | 7.54 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr4_-_42856771 | 7.31 |
ENSMUST00000107981.2
|
Gm12394
|
predicted gene 12394 |
| chr14_-_52151537 | 7.01 |
ENSMUST00000227402.2
ENSMUST00000227237.2 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr13_-_113800172 | 6.86 |
ENSMUST00000054650.5
|
Hspb3
|
heat shock protein 3 |
| chr5_+_25964985 | 6.82 |
ENSMUST00000128727.8
ENSMUST00000088244.6 |
Actr3b
|
ARP3 actin-related protein 3B |
| chr15_+_101152078 | 6.75 |
ENSMUST00000228985.2
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr2_+_136555364 | 6.66 |
ENSMUST00000028727.11
ENSMUST00000110098.4 |
Snap25
|
synaptosomal-associated protein 25 |
| chr15_+_101164719 | 6.36 |
ENSMUST00000230814.2
ENSMUST00000023779.8 |
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr8_+_31579499 | 6.30 |
ENSMUST00000036631.14
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr3_-_122828592 | 6.28 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr14_-_52151026 | 6.25 |
ENSMUST00000228164.2
|
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr14_+_26616514 | 6.11 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr3_+_68491487 | 6.05 |
ENSMUST00000182997.3
|
Schip1
|
schwannomin interacting protein 1 |
| chr1_-_66974492 | 6.05 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr3_+_32871669 | 5.85 |
ENSMUST00000072312.12
ENSMUST00000108228.8 |
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr12_-_40249314 | 5.79 |
ENSMUST00000095760.3
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr8_+_31579633 | 5.72 |
ENSMUST00000170204.8
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr16_-_28571820 | 5.60 |
ENSMUST00000232352.2
|
Fgf12
|
fibroblast growth factor 12 |
| chr7_+_44984681 | 5.59 |
ENSMUST00000085351.7
|
Hrc
|
histidine rich calcium binding protein |
| chr8_+_15107646 | 5.50 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr11_+_67689094 | 5.49 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr14_-_51134930 | 5.45 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr6_+_42263609 | 5.32 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr17_-_26056056 | 5.23 |
ENSMUST00000183929.8
ENSMUST00000184865.2 ENSMUST00000026831.14 |
Rhbdl1
|
rhomboid like 1 |
| chr14_-_20706556 | 5.13 |
ENSMUST00000090469.8
|
Myoz1
|
myozenin 1 |
| chr12_-_40249489 | 5.12 |
ENSMUST00000220951.2
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr1_-_66974694 | 5.09 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr10_-_70428611 | 5.00 |
ENSMUST00000162251.8
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr11_+_54195006 | 5.00 |
ENSMUST00000108904.10
ENSMUST00000108905.10 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr7_-_25374472 | 4.91 |
ENSMUST00000108404.8
ENSMUST00000108405.2 ENSMUST00000079439.10 |
Tmem91
|
transmembrane protein 91 |
| chr7_-_142453722 | 4.85 |
ENSMUST00000000219.10
|
Th
|
tyrosine hydroxylase |
| chr4_-_73869071 | 4.73 |
ENSMUST00000095023.2
ENSMUST00000030101.4 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr3_+_146558114 | 4.72 |
ENSMUST00000170055.8
ENSMUST00000037942.11 |
Ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr1_+_9618173 | 4.60 |
ENSMUST00000144177.8
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr3_-_158267771 | 4.59 |
ENSMUST00000199890.5
ENSMUST00000238317.3 ENSMUST00000200137.5 ENSMUST00000106044.6 |
Lrrc7
|
leucine rich repeat containing 7 |
| chrX_+_156481906 | 4.58 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr12_+_69288606 | 4.58 |
ENSMUST00000063445.13
|
Klhdc1
|
kelch domain containing 1 |
| chr2_+_69500444 | 4.55 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr14_-_51134906 | 4.47 |
ENSMUST00000170855.2
|
Klhl33
|
kelch-like 33 |
| chr3_+_96503944 | 4.35 |
ENSMUST00000058943.8
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr15_+_98065039 | 4.32 |
ENSMUST00000031914.6
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr7_+_80707328 | 4.31 |
ENSMUST00000107348.2
|
Alpk3
|
alpha-kinase 3 |
| chr2_-_17395765 | 4.29 |
ENSMUST00000177966.2
|
Nebl
|
nebulette |
| chr2_+_90948481 | 4.23 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr7_-_105230479 | 4.07 |
ENSMUST00000191601.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr10_+_32959472 | 4.01 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chrX_+_163221035 | 3.99 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr10_-_88440869 | 3.87 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr11_+_54194831 | 3.85 |
ENSMUST00000000145.12
ENSMUST00000138515.8 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr2_+_120294046 | 3.84 |
ENSMUST00000028749.15
ENSMUST00000110721.9 ENSMUST00000239364.2 |
Capn3
|
calpain 3 |
| chr8_+_23548541 | 3.84 |
ENSMUST00000173248.8
|
Ank1
|
ankyrin 1, erythroid |
| chrY_-_1286623 | 3.76 |
ENSMUST00000091190.12
|
Ddx3y
|
DEAD box helicase 3, Y-linked |
| chr6_+_42263644 | 3.75 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr5_+_66903174 | 3.70 |
ENSMUST00000101164.11
ENSMUST00000238785.2 |
Limch1
|
LIM and calponin homology domains 1 |
| chr3_-_113368407 | 3.69 |
ENSMUST00000106540.8
|
Amy1
|
amylase 1, salivary |
| chr17_+_34258411 | 3.68 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr18_-_25886908 | 3.65 |
ENSMUST00000115816.3
ENSMUST00000223704.2 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr7_+_140343652 | 3.64 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr14_+_55813074 | 3.62 |
ENSMUST00000022826.7
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr5_+_122239007 | 3.61 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr1_-_172156884 | 3.61 |
ENSMUST00000062387.8
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr7_+_90739904 | 3.58 |
ENSMUST00000107196.10
ENSMUST00000074273.10 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr1_-_172047282 | 3.58 |
ENSMUST00000170700.2
ENSMUST00000003554.11 |
Casq1
|
calsequestrin 1 |
| chr2_-_79738773 | 3.53 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr4_+_156300325 | 3.53 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr7_+_141996067 | 3.53 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr7_-_101749433 | 3.52 |
ENSMUST00000106937.8
|
Art5
|
ADP-ribosyltransferase 5 |
| chr6_-_55109960 | 3.51 |
ENSMUST00000003568.15
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr5_+_122239030 | 3.45 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr15_+_73620213 | 3.33 |
ENSMUST00000053232.8
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr6_-_112924205 | 3.23 |
ENSMUST00000088373.11
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr7_+_81512421 | 3.19 |
ENSMUST00000119543.2
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr5_-_116560916 | 3.19 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr11_+_73051228 | 3.19 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr17_+_12597490 | 3.16 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr1_-_80642969 | 3.14 |
ENSMUST00000190983.7
ENSMUST00000191449.2 |
Dock10
|
dedicator of cytokinesis 10 |
| chr2_-_79738734 | 3.12 |
ENSMUST00000090756.11
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_+_67276338 | 3.11 |
ENSMUST00000239060.2
ENSMUST00000028410.4 ENSMUST00000112347.8 ENSMUST00000238878.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr18_-_3299536 | 2.97 |
ENSMUST00000129435.8
ENSMUST00000122958.8 |
Crem
|
cAMP responsive element modulator |
| chr1_+_75351914 | 2.97 |
ENSMUST00000087122.12
|
Speg
|
SPEG complex locus |
| chr8_-_106670014 | 2.96 |
ENSMUST00000038896.8
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr6_-_112923715 | 2.96 |
ENSMUST00000113169.9
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr1_-_80643024 | 2.87 |
ENSMUST00000187774.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr18_+_67597929 | 2.86 |
ENSMUST00000025411.9
|
Prelid3a
|
PRELI domain containing 3A |
| chr18_-_25887173 | 2.84 |
ENSMUST00000225477.2
|
Celf4
|
CUGBP, Elav-like family member 4 |
| chr6_+_48566540 | 2.81 |
ENSMUST00000204521.2
|
Gm45021
|
predicted gene 45021 |
| chr7_-_48497771 | 2.76 |
ENSMUST00000032658.14
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr13_-_74512111 | 2.76 |
ENSMUST00000022064.5
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chr8_-_84169107 | 2.71 |
ENSMUST00000212031.2
ENSMUST00000167525.3 |
Scoc
|
short coiled-coil protein |
| chr2_-_171885386 | 2.71 |
ENSMUST00000087950.4
|
Cbln4
|
cerebellin 4 precursor protein |
| chr1_-_172156828 | 2.70 |
ENSMUST00000194204.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr11_-_94867153 | 2.67 |
ENSMUST00000103162.8
ENSMUST00000166320.8 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr7_-_105230395 | 2.65 |
ENSMUST00000188726.2
ENSMUST00000188440.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr11_+_104468107 | 2.64 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr9_+_102382949 | 2.64 |
ENSMUST00000039390.6
|
Ky
|
kyphoscoliosis peptidase |
| chr10_+_90412827 | 2.63 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_-_88604404 | 2.60 |
ENSMUST00000120933.5
|
Kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr7_+_127603083 | 2.58 |
ENSMUST00000106248.8
|
Trim72
|
tripartite motif-containing 72 |
| chrX_+_156482116 | 2.58 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr11_+_54194624 | 2.56 |
ENSMUST00000093106.12
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr15_-_89310060 | 2.48 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr7_+_15863679 | 2.46 |
ENSMUST00000211649.2
|
Slc8a2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr9_-_54554483 | 2.37 |
ENSMUST00000128163.8
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr10_-_18619658 | 2.30 |
ENSMUST00000215836.2
|
Arfgef3
|
ARFGEF family member 3 |
| chr6_-_128503666 | 2.29 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr11_+_70548513 | 2.28 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle |
| chr11_+_70548022 | 2.27 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr1_-_24626492 | 2.27 |
ENSMUST00000051344.6
ENSMUST00000115244.9 |
Col19a1
|
collagen, type XIX, alpha 1 |
| chr7_+_29821340 | 2.27 |
ENSMUST00000098596.11
ENSMUST00000153792.2 |
Zfp382
|
zinc finger protein 382 |
| chr12_-_84993695 | 2.26 |
ENSMUST00000169161.8
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr11_+_105069591 | 2.26 |
ENSMUST00000106939.9
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr11_+_104467791 | 2.25 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chr2_+_91086489 | 2.18 |
ENSMUST00000154959.8
ENSMUST00000059566.11 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr17_-_33049169 | 2.14 |
ENSMUST00000201499.2
ENSMUST00000201876.4 ENSMUST00000202759.2 ENSMUST00000202988.4 |
Zfp799
|
zinc finger protein 799 |
| chr14_-_20718337 | 2.10 |
ENSMUST00000057090.12
ENSMUST00000117386.2 |
Synpo2l
|
synaptopodin 2-like |
| chr1_-_75195127 | 2.08 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr5_+_66903195 | 2.06 |
ENSMUST00000118242.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr15_-_102630589 | 2.06 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr7_+_120276801 | 2.02 |
ENSMUST00000208454.2
ENSMUST00000060175.8 |
Mosmo
|
modulator of smoothened |
| chr15_-_89309998 | 2.00 |
ENSMUST00000168376.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr2_+_118493713 | 1.98 |
ENSMUST00000099557.10
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr1_+_36411312 | 1.98 |
ENSMUST00000179162.7
|
Fer1l5
|
fer-1-like 5 (C. elegans) |
| chr14_+_29700319 | 1.91 |
ENSMUST00000224797.2
|
Actr8
|
ARP8 actin-related protein 8 |
| chr19_-_46958001 | 1.81 |
ENSMUST00000235234.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr6_-_68857658 | 1.79 |
ENSMUST00000198756.2
|
Gm42543
|
predicted gene 42543 |
| chr9_+_110948492 | 1.79 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr14_+_29700294 | 1.76 |
ENSMUST00000016115.6
|
Actr8
|
ARP8 actin-related protein 8 |
| chr6_+_121277186 | 1.75 |
ENSMUST00000064580.14
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr2_+_43638814 | 1.74 |
ENSMUST00000112824.8
ENSMUST00000055776.8 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr6_-_124942366 | 1.72 |
ENSMUST00000129446.8
ENSMUST00000032220.15 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr18_+_37093321 | 1.69 |
ENSMUST00000192168.2
|
Pcdha5
|
protocadherin alpha 5 |
| chr10_-_117074501 | 1.67 |
ENSMUST00000159193.8
ENSMUST00000020392.5 |
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
| chrX_-_94521712 | 1.66 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr4_+_116008623 | 1.65 |
ENSMUST00000106494.3
|
Pomgnt1
|
protein O-linked mannose beta 1,2-N-acetylglucosaminyltransferase |
| chr2_-_160169414 | 1.62 |
ENSMUST00000099127.3
|
Gm826
|
predicted gene 826 |
| chr7_+_141995545 | 1.61 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr18_-_3281089 | 1.61 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chrX_-_63320543 | 1.58 |
ENSMUST00000114679.2
ENSMUST00000069926.14 |
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr8_+_95584146 | 1.57 |
ENSMUST00000211939.2
ENSMUST00000212124.2 |
Polr2c
|
polymerase (RNA) II (DNA directed) polypeptide C |
| chr19_-_37153436 | 1.57 |
ENSMUST00000142973.2
ENSMUST00000154376.8 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr5_-_3943907 | 1.55 |
ENSMUST00000117463.2
ENSMUST00000044746.5 |
Mterf1a
|
mitochondrial transcription termination factor 1a |
| chr11_+_102159558 | 1.52 |
ENSMUST00000036467.5
|
Asb16
|
ankyrin repeat and SOCS box-containing 16 |
| chr6_+_137387718 | 1.52 |
ENSMUST00000167002.4
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr1_-_173707677 | 1.52 |
ENSMUST00000190651.4
ENSMUST00000188804.7 |
Mndal
|
myeloid nuclear differentiation antigen like |
| chr5_+_120614587 | 1.44 |
ENSMUST00000201684.4
ENSMUST00000066540.14 |
Sds
|
serine dehydratase |
| chr19_-_40175709 | 1.42 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr9_+_74959259 | 1.42 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr13_+_89687915 | 1.41 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr10_-_39775182 | 1.40 |
ENSMUST00000178045.9
ENSMUST00000178563.3 |
Mfsd4b4
|
major facilitator superfamily domain containing 4B4 |
| chr1_-_39844467 | 1.38 |
ENSMUST00000171319.4
|
Gm3646
|
predicted gene 3646 |
| chr6_-_124942457 | 1.37 |
ENSMUST00000112439.9
|
Cops7a
|
COP9 signalosome subunit 7A |
| chr18_+_60426444 | 1.34 |
ENSMUST00000171297.2
|
F830016B08Rik
|
RIKEN cDNA F830016B08 gene |
| chr1_-_14374794 | 1.34 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_+_155433858 | 1.32 |
ENSMUST00000080138.13
ENSMUST00000035560.9 ENSMUST00000097529.5 |
Acbd6
|
acyl-Coenzyme A binding domain containing 6 |
| chr3_-_154302679 | 1.31 |
ENSMUST00000052774.8
ENSMUST00000170461.8 ENSMUST00000122976.2 |
Tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr4_-_115911053 | 1.29 |
ENSMUST00000030475.3
|
Nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr3_+_101993731 | 1.28 |
ENSMUST00000029454.12
|
Casq2
|
calsequestrin 2 |
| chr13_-_23553327 | 1.28 |
ENSMUST00000125328.2
ENSMUST00000145451.8 ENSMUST00000050101.9 |
Zfp322a
|
zinc finger protein 322A |
| chr18_-_3299452 | 1.24 |
ENSMUST00000126578.8
|
Crem
|
cAMP responsive element modulator |
| chr18_+_37898633 | 1.23 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr3_-_154760978 | 1.22 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr14_-_21764638 | 1.22 |
ENSMUST00000073870.7
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr1_-_160958998 | 1.18 |
ENSMUST00000111611.8
|
Klhl20
|
kelch-like 20 |
| chr9_-_110819639 | 1.17 |
ENSMUST00000198702.2
|
Rtp3
|
receptor transporter protein 3 |
| chr13_+_83672389 | 1.16 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr9_-_119852624 | 1.16 |
ENSMUST00000111635.4
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr5_-_24652775 | 1.14 |
ENSMUST00000123167.2
ENSMUST00000030799.15 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr16_-_17019352 | 1.13 |
ENSMUST00000090192.12
|
Ube2l3
|
ubiquitin-conjugating enzyme E2L 3 |
| chr12_-_65010124 | 1.13 |
ENSMUST00000021331.9
|
Klhl28
|
kelch-like 28 |
| chr7_-_12552764 | 1.12 |
ENSMUST00000108546.2
ENSMUST00000072222.8 |
Zfp329
|
zinc finger protein 329 |
| chr18_-_80243812 | 1.12 |
ENSMUST00000025462.7
|
Rbfa
|
ribosome binding factor A |
| chr2_-_111779785 | 1.11 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr7_+_27878894 | 1.11 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr5_+_21990251 | 1.10 |
ENSMUST00000239497.2
ENSMUST00000030769.7 |
Psmc2
|
proteasome (prosome, macropain) 26S subunit, ATPase 2 |
| chr3_-_20209220 | 1.09 |
ENSMUST00000184552.2
|
Gyg
|
glycogenin |
| chr6_-_35516700 | 1.09 |
ENSMUST00000201026.2
ENSMUST00000031866.9 |
Mtpn
|
myotrophin |
| chr3_+_101993787 | 1.08 |
ENSMUST00000165540.9
ENSMUST00000164123.2 |
Casq2
|
calsequestrin 2 |
| chr13_-_67229566 | 1.06 |
ENSMUST00000109742.11
ENSMUST00000190566.3 |
Zfp708
|
zinc finger protein 708 |
| chr14_+_47886748 | 1.01 |
ENSMUST00000190252.7
ENSMUST00000186466.3 |
Ktn1
|
kinectin 1 |
| chr12_-_34956910 | 1.00 |
ENSMUST00000239321.2
|
Hdac9
|
histone deacetylase 9 |
| chr18_+_37646674 | 0.98 |
ENSMUST00000061405.6
|
Pcdhb21
|
protocadherin beta 21 |
| chr13_-_23553139 | 0.96 |
ENSMUST00000152557.8
|
Zfp322a
|
zinc finger protein 322A |
| chr2_+_25572737 | 0.95 |
ENSMUST00000058912.3
|
Lcn10
|
lipocalin 10 |
| chr2_-_85966272 | 0.95 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr8_-_112718888 | 0.94 |
ENSMUST00000034427.12
ENSMUST00000139820.8 |
Adat1
|
adenosine deaminase, tRNA-specific 1 |
| chr6_+_137387729 | 0.93 |
ENSMUST00000203914.2
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr16_-_58930996 | 0.93 |
ENSMUST00000076262.4
|
Olfr193
|
olfactory receptor 193 |
| chr9_-_19799300 | 0.93 |
ENSMUST00000079660.5
|
Olfr862
|
olfactory receptor 862 |
| chr1_-_173770010 | 0.93 |
ENSMUST00000042228.15
ENSMUST00000081216.12 ENSMUST00000129829.8 ENSMUST00000123708.8 |
Ifi203
|
interferon activated gene 203 |
| chr1_+_15382676 | 0.93 |
ENSMUST00000170146.3
|
Kcnb2
|
potassium voltage gated channel, Shab-related subfamily, member 2 |
| chr5_+_20112704 | 0.92 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 12.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 2.4 | 7.1 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 2.3 | 11.4 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 2.2 | 6.7 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 2.2 | 13.3 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 2.1 | 8.4 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 1.6 | 13.0 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.5 | 21.3 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 1.5 | 5.9 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 1.4 | 5.6 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 1.4 | 9.5 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 1.1 | 5.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.1 | 6.7 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 1.1 | 10.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.0 | 4.9 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 1.0 | 3.8 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.9 | 2.8 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.9 | 8.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.9 | 3.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.9 | 3.5 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.7 | 3.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.7 | 4.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.6 | 9.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.6 | 3.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.6 | 9.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.6 | 2.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.5 | 1.6 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.5 | 1.5 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.5 | 6.5 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.5 | 3.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 | 6.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.4 | 9.1 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.4 | 4.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.4 | 1.8 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.4 | 2.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 6.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 1.5 | GO:0036343 | psychomotor behavior(GO:0036343) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.3 | 4.5 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.3 | 2.4 | GO:0071313 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) cellular response to caffeine(GO:0071313) |
| 0.3 | 2.3 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.3 | 2.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 3.0 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.2 | 2.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 5.5 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.2 | 6.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 0.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.6 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.2 | 2.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 3.5 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 0.4 | GO:0003169 | coronary vein morphogenesis(GO:0003169) |
| 0.2 | 3.6 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.2 | 3.3 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.2 | 1.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.2 | 9.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.2 | 2.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 0.8 | GO:0090669 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.2 | 3.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 2.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 5.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.7 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.3 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.1 | 1.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 10.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 0.4 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 1.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 30.0 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 6.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 6.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.6 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.3 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 2.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.7 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.1 | 0.4 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.1 | 0.3 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 6.4 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 1.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 3.8 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.2 | GO:2000424 | T-helper 1 cell lineage commitment(GO:0002296) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 1.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.7 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 3.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 3.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 5.6 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 2.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 0.2 | GO:0048007 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.3 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 2.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 2.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 4.5 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 1.4 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.6 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 1.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.4 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 1.6 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 10.4 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 4.6 | GO:0006818 | hydrogen transport(GO:0006818) |
| 0.0 | 2.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 5.7 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 2.6 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 1.9 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 1.2 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 1.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 2.9 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.3 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.5 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 1.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 3.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.8 | GO:0070229 | negative regulation of lymphocyte apoptotic process(GO:0070229) |
| 0.0 | 0.4 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.8 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 1.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 4.0 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 1.6 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 2.3 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.9 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 2.3 | GO:0030198 | extracellular matrix organization(GO:0030198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 21.3 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 2.0 | 5.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.8 | 5.5 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 1.4 | 4.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 1.3 | 6.7 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.0 | 4.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.9 | 7.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.6 | 6.7 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.6 | 4.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.5 | 3.2 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.5 | 9.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 5.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 7.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 28.1 | GO:0031672 | A band(GO:0031672) |
| 0.3 | 6.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 4.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 2.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 9.5 | GO:0036379 | myofilament(GO:0036379) |
| 0.3 | 5.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 0.8 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 3.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 4.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 3.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 3.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 11.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 1.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 28.3 | GO:0030016 | myofibril(GO:0030016) |
| 0.1 | 3.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 8.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 24.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 2.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 11.3 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 12.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 4.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 1.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 8.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.2 | GO:0005672 | RNA polymerase I transcription factor complex(GO:0000120) transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 4.6 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 5.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 3.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 6.8 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 4.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 3.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 21.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0044297 | cell body(GO:0044297) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 2.3 | 11.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 2.0 | 12.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 1.3 | 21.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.2 | 4.9 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 1.2 | 4.7 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 1.2 | 5.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.2 | 3.5 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.9 | 3.5 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.8 | 4.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.8 | 8.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.7 | 4.5 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.7 | 6.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.7 | 2.8 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.7 | 13.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.6 | 1.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.6 | 6.6 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.6 | 1.7 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.5 | 3.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.5 | 10.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 5.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.5 | 3.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.5 | 1.4 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.5 | 2.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.5 | 9.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 1.3 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.4 | 9.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.4 | 3.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 3.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 9.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 2.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.3 | 4.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 3.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 5.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 10.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 2.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 12.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.2 | 3.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 6.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 2.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 0.6 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.2 | 0.6 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.2 | 6.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 2.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 1.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.2 | 2.3 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.2 | 1.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 1.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 2.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 3.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 5.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.6 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.7 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 5.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 6.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 6.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 8.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 5.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 1.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 3.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 2.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:0001129 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 3.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 31.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.2 | GO:0035198 | AT DNA binding(GO:0003680) miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 1.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 6.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 4.0 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 1.6 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 1.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 23.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 7.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 10.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 6.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 5.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 11.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 6.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 10.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 6.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 6.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.6 | 41.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 21.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.5 | 6.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 11.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 13.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 4.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 6.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 4.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 8.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 8.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 6.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 4.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 6.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 9.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 11.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 6.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 1.2 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |