Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Meis1
Z-value: 0.80
Transcription factors associated with Meis1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meis1
|
ENSMUSG00000020160.19 | Meis1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meis1 | mm39_v1_chr11_-_18968955_18968973 | 0.53 | 1.4e-06 | Click! |
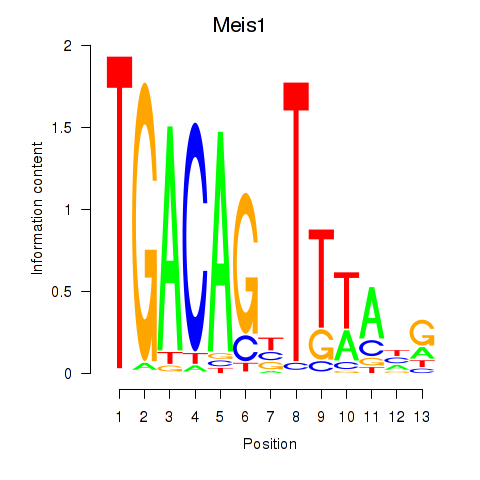
Activity profile of Meis1 motif
Sorted Z-values of Meis1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Meis1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_41098174 | 6.43 |
ENSMUST00000055327.8
|
Aqp3
|
aquaporin 3 |
| chr3_-_92627651 | 5.59 |
ENSMUST00000047153.4
|
Lce1f
|
late cornified envelope 1F |
| chr1_-_107206091 | 5.26 |
ENSMUST00000166100.2
ENSMUST00000027565.5 |
Serpinb3b
Serpinb3c
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3B serine (or cysteine) peptidase inhibitor, clade B, member 3C |
| chr13_+_49761506 | 4.88 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr9_-_26717686 | 4.82 |
ENSMUST00000162702.8
ENSMUST00000040398.14 ENSMUST00000066560.13 |
Glb1l2
|
galactosidase, beta 1-like 2 |
| chr3_-_10273628 | 4.00 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr15_+_74586682 | 3.98 |
ENSMUST00000023265.5
|
Psca
|
prostate stem cell antigen |
| chr6_-_133830613 | 3.85 |
ENSMUST00000203168.2
ENSMUST00000048032.5 |
Kap
|
kidney androgen regulated protein |
| chr13_+_49697919 | 3.82 |
ENSMUST00000177948.2
ENSMUST00000021820.14 |
Aspn
|
asporin |
| chr7_+_28563255 | 3.49 |
ENSMUST00000138272.8
|
Lgals7
|
lectin, galactose binding, soluble 7 |
| chr10_-_84938350 | 3.12 |
ENSMUST00000059383.8
ENSMUST00000216889.2 |
Fhl4
|
four and a half LIM domains 4 |
| chr2_-_85349362 | 3.11 |
ENSMUST00000099923.2
|
Fads2b
|
fatty acid desaturase 2B |
| chr8_+_127790772 | 3.06 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr9_+_44045859 | 2.98 |
ENSMUST00000034650.15
ENSMUST00000098852.3 ENSMUST00000216002.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr2_-_60503998 | 2.70 |
ENSMUST00000059888.15
ENSMUST00000154764.2 |
Itgb6
|
integrin beta 6 |
| chr7_-_89176294 | 2.67 |
ENSMUST00000207932.2
|
Prss23
|
protease, serine 23 |
| chr14_+_103750763 | 2.62 |
ENSMUST00000227322.2
|
Scel
|
sciellin |
| chr16_-_92156312 | 2.59 |
ENSMUST00000051705.7
|
Kcne1
|
potassium voltage-gated channel, Isk-related subfamily, member 1 |
| chr12_+_119278005 | 2.51 |
ENSMUST00000222784.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr15_-_34679321 | 2.50 |
ENSMUST00000040791.9
|
Nipal2
|
NIPA-like domain containing 2 |
| chr1_-_173054760 | 2.45 |
ENSMUST00000193017.2
ENSMUST00000049706.11 |
Fcer1a
|
Fc receptor, IgE, high affinity I, alpha polypeptide |
| chr1_-_15962881 | 2.11 |
ENSMUST00000040695.5
|
Sbspon
|
somatomedin B and thrombospondin, type 1 domain containing |
| chr9_-_107483855 | 2.04 |
ENSMUST00000073448.12
ENSMUST00000194606.2 ENSMUST00000195662.6 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr17_-_24877431 | 1.93 |
ENSMUST00000095544.6
|
Npw
|
neuropeptide W |
| chr7_+_45084257 | 1.87 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr9_+_107173907 | 1.77 |
ENSMUST00000168260.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr18_+_44237577 | 1.75 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr3_-_87170903 | 1.73 |
ENSMUST00000090986.11
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr18_+_44237474 | 1.71 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr9_-_71499628 | 1.69 |
ENSMUST00000093823.8
|
Myzap
|
myocardial zonula adherens protein |
| chr1_+_130659700 | 1.68 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr15_-_74869483 | 1.66 |
ENSMUST00000023248.13
|
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr15_-_74869684 | 1.63 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr19_-_11261177 | 1.59 |
ENSMUST00000186937.7
ENSMUST00000067673.13 |
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr7_-_19133783 | 1.58 |
ENSMUST00000047170.10
ENSMUST00000108459.9 |
Klc3
|
kinesin light chain 3 |
| chr4_+_19818718 | 1.54 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr19_-_20704896 | 1.52 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr3_+_157272504 | 1.51 |
ENSMUST00000041175.13
ENSMUST00000173533.2 |
Ptger3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr12_+_75355082 | 1.51 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr8_-_71219299 | 1.48 |
ENSMUST00000222087.4
|
Ifi30
|
interferon gamma inducible protein 30 |
| chr6_-_4747157 | 1.47 |
ENSMUST00000126151.8
ENSMUST00000115577.9 ENSMUST00000101677.9 ENSMUST00000115579.8 ENSMUST00000004750.15 |
Sgce
|
sarcoglycan, epsilon |
| chr19_+_4008645 | 1.44 |
ENSMUST00000179433.8
|
Aldh3b3
|
aldehyde dehydrogenase 3 family, member B3 |
| chr6_-_4747066 | 1.41 |
ENSMUST00000090686.11
ENSMUST00000133306.8 |
Sgce
|
sarcoglycan, epsilon |
| chr2_-_65069383 | 1.40 |
ENSMUST00000155916.8
ENSMUST00000156643.2 |
Cobll1
|
Cobl-like 1 |
| chr9_+_107174081 | 1.40 |
ENSMUST00000167072.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr10_+_69044376 | 1.38 |
ENSMUST00000164034.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr7_-_144232586 | 1.37 |
ENSMUST00000131731.2
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr5_+_16139760 | 1.33 |
ENSMUST00000101581.10
ENSMUST00000039370.14 ENSMUST00000199704.5 ENSMUST00000180204.8 ENSMUST00000078272.13 ENSMUST00000115281.7 |
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr6_+_65929546 | 1.33 |
ENSMUST00000043382.9
|
4930544G11Rik
|
RIKEN cDNA 4930544G11 gene |
| chr12_-_108668520 | 1.31 |
ENSMUST00000167978.9
ENSMUST00000021691.6 |
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr7_+_12661337 | 1.31 |
ENSMUST00000045870.5
|
Rnf225
|
ring finger protein 225 |
| chr8_+_129450766 | 1.27 |
ENSMUST00000149116.2
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta) |
| chr10_+_56255184 | 1.27 |
ENSMUST00000220069.2
|
Gja1
|
gap junction protein, alpha 1 |
| chr2_+_144665576 | 1.26 |
ENSMUST00000028918.4
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr9_+_53295317 | 1.20 |
ENSMUST00000037853.5
|
Poglut3
|
protein O-glucosyltransferase 3 |
| chr11_+_80749184 | 1.19 |
ENSMUST00000103223.8
ENSMUST00000103222.4 |
Spaca3
|
sperm acrosome associated 3 |
| chr1_-_153061758 | 1.15 |
ENSMUST00000185356.7
|
Lamc2
|
laminin, gamma 2 |
| chr15_-_26895660 | 1.15 |
ENSMUST00000059204.11
|
Fbxl7
|
F-box and leucine-rich repeat protein 7 |
| chr19_-_40371016 | 1.14 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr17_-_65946817 | 1.14 |
ENSMUST00000233702.2
|
Txndc2
|
thioredoxin domain containing 2 (spermatozoa) |
| chrX_-_72703330 | 1.13 |
ENSMUST00000114473.8
ENSMUST00000002087.14 |
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr19_-_28657477 | 1.11 |
ENSMUST00000162022.8
ENSMUST00000112612.9 |
Glis3
|
GLIS family zinc finger 3 |
| chr3_-_87171005 | 1.11 |
ENSMUST00000146512.2
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr1_-_56676589 | 1.11 |
ENSMUST00000062085.6
|
Hsfy2
|
heat shock transcription factor, Y-linked 2 |
| chr6_-_124613044 | 1.11 |
ENSMUST00000068797.3
ENSMUST00000218020.2 |
C1s2
|
complement component 1, s subcomponent 2 |
| chr1_-_66974694 | 1.11 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr2_-_25126701 | 1.10 |
ENSMUST00000205192.2
ENSMUST00000091318.5 |
Rnf224
|
ring finger protein 224 |
| chr7_+_142025575 | 1.10 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr8_-_83128437 | 1.09 |
ENSMUST00000209573.3
|
Il15
|
interleukin 15 |
| chr19_+_42135812 | 1.07 |
ENSMUST00000061111.10
|
Marveld1
|
MARVEL (membrane-associating) domain containing 1 |
| chr2_-_179931672 | 1.05 |
ENSMUST00000038529.2
|
Rbbp8nl
|
RBBP8 N-terminal like |
| chr5_+_16139683 | 1.05 |
ENSMUST00000167946.9
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr10_-_14420725 | 1.02 |
ENSMUST00000041168.6
ENSMUST00000238680.2 |
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr3_+_109030419 | 1.02 |
ENSMUST00000029477.11
|
Slc25a24
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 24 |
| chr6_+_30509826 | 1.01 |
ENSMUST00000031797.11
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr17_+_26895344 | 1.00 |
ENSMUST00000015719.16
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr1_-_66974492 | 1.00 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr10_-_128329693 | 0.99 |
ENSMUST00000217776.2
ENSMUST00000219236.2 ENSMUST00000217733.2 ENSMUST00000218127.2 ENSMUST00000217969.2 ENSMUST00000164181.2 |
Myl6
|
myosin, light polypeptide 6, alkali, smooth muscle and non-muscle |
| chr16_+_48692976 | 0.99 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr17_+_75742881 | 0.98 |
ENSMUST00000164192.9
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr2_-_3420102 | 0.98 |
ENSMUST00000115082.10
|
Meig1
|
meiosis expressed gene 1 |
| chr19_+_47854206 | 0.97 |
ENSMUST00000056159.11
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr4_-_87724512 | 0.97 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr16_-_26345493 | 0.96 |
ENSMUST00000165687.3
|
Tmem207
|
transmembrane protein 207 |
| chr7_+_142025817 | 0.94 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr15_+_100202079 | 0.91 |
ENSMUST00000230252.2
ENSMUST00000231166.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr17_+_25527616 | 0.90 |
ENSMUST00000015267.5
|
Prss28
|
protease, serine 28 |
| chr15_+_100202061 | 0.88 |
ENSMUST00000229574.2
ENSMUST00000229217.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr15_+_34453432 | 0.88 |
ENSMUST00000060894.9
|
Erich5
|
glutamate rich 5 |
| chr16_+_90623630 | 0.87 |
ENSMUST00000099548.10
|
Eva1c
|
eva-1 homolog C (C. elegans) |
| chr9_-_107863062 | 0.86 |
ENSMUST00000048568.6
|
Inka1
|
inka box actin regulator 1 |
| chr2_-_142743354 | 0.85 |
ENSMUST00000211861.2
|
Kif16b
|
kinesin family member 16B |
| chr7_+_44221791 | 0.85 |
ENSMUST00000002274.10
|
Napsa
|
napsin A aspartic peptidase |
| chr6_-_131655849 | 0.84 |
ENSMUST00000076756.3
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chr8_-_3517617 | 0.82 |
ENSMUST00000111081.10
ENSMUST00000004686.13 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr6_+_116627567 | 0.82 |
ENSMUST00000067354.10
ENSMUST00000178241.4 |
Depp1
|
DEPP1 autophagy regulator |
| chr5_+_16139909 | 0.81 |
ENSMUST00000196750.2
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr5_-_115257336 | 0.79 |
ENSMUST00000031524.11
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain |
| chr9_-_14682300 | 0.78 |
ENSMUST00000191047.7
ENSMUST00000060330.5 |
1700012B09Rik
|
RIKEN cDNA 1700012B09 gene |
| chrX_+_142936691 | 0.77 |
ENSMUST00000135687.2
|
A730046J19Rik
|
RIKEN cDNA A730046J19 gene |
| chr19_+_47854249 | 0.76 |
ENSMUST00000238084.2
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr12_+_91367764 | 0.76 |
ENSMUST00000021346.14
ENSMUST00000021343.8 |
Tshr
|
thyroid stimulating hormone receptor |
| chr8_+_109441276 | 0.76 |
ENSMUST00000043896.10
|
Zfhx3
|
zinc finger homeobox 3 |
| chr9_-_104214899 | 0.74 |
ENSMUST00000112590.3
|
Acpp
|
acid phosphatase, prostate |
| chrX_-_105884178 | 0.74 |
ENSMUST00000062010.10
|
Rtl3
|
retrotransposon Gag like 3 |
| chr2_-_17465410 | 0.74 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr6_-_54941673 | 0.74 |
ENSMUST00000203837.2
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr2_+_25293056 | 0.73 |
ENSMUST00000071442.12
|
Npdc1
|
neural proliferation, differentiation and control 1 |
| chr16_-_95260104 | 0.73 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr5_-_151351628 | 0.72 |
ENSMUST00000202365.2
ENSMUST00000186838.2 |
Gm42906
D730045B01Rik
|
predicted gene 42906 RIKEN cDNA D730045B01 gene |
| chr11_+_66915969 | 0.72 |
ENSMUST00000079077.12
ENSMUST00000061786.6 |
Tmem220
|
transmembrane protein 220 |
| chr10_+_81464536 | 0.72 |
ENSMUST00000129622.2
|
Ankrd24
|
ankyrin repeat domain 24 |
| chr7_+_78563150 | 0.72 |
ENSMUST00000118867.8
|
Isg20
|
interferon-stimulated protein |
| chr12_-_24731768 | 0.71 |
ENSMUST00000189849.7
ENSMUST00000186602.7 |
Cys1
|
cystin 1 |
| chr8_-_8711211 | 0.71 |
ENSMUST00000001319.15
|
Efnb2
|
ephrin B2 |
| chr8_+_48277493 | 0.70 |
ENSMUST00000038693.8
|
Cldn22
|
claudin 22 |
| chr4_+_52607204 | 0.70 |
ENSMUST00000107671.2
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr5_+_114142842 | 0.69 |
ENSMUST00000161610.6
|
Dao
|
D-amino acid oxidase |
| chr16_-_38533597 | 0.68 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr9_+_106325828 | 0.67 |
ENSMUST00000217496.2
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr3_-_89294430 | 0.67 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr2_+_88508201 | 0.67 |
ENSMUST00000081697.2
|
Olfr1193
|
olfactory receptor 1193 |
| chr15_-_103338814 | 0.66 |
ENSMUST00000147389.8
ENSMUST00000023129.15 |
Gtsf1
|
gametocyte specific factor 1 |
| chr4_-_87724533 | 0.65 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr6_-_118116062 | 0.65 |
ENSMUST00000049344.15
|
Csgalnact2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr15_+_100202021 | 0.64 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr2_-_30095805 | 0.64 |
ENSMUST00000113663.9
ENSMUST00000044038.10 |
Kyat1
|
kynurenine aminotransferase 1 |
| chr6_+_34389269 | 0.64 |
ENSMUST00000007449.9
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr12_-_50695881 | 0.64 |
ENSMUST00000002765.9
|
Prkd1
|
protein kinase D1 |
| chr4_+_39450265 | 0.64 |
ENSMUST00000029955.5
|
1700009N14Rik
|
RIKEN cDNA 1700009N14 gene |
| chr5_+_136086045 | 0.64 |
ENSMUST00000111137.8
ENSMUST00000156530.8 ENSMUST00000006303.11 |
Upk3bl
|
uroplakin 3B-like |
| chr6_+_29060215 | 0.63 |
ENSMUST00000069789.12
|
Lep
|
leptin |
| chr13_+_38529062 | 0.63 |
ENSMUST00000171970.3
|
Bmp6
|
bone morphogenetic protein 6 |
| chr11_-_100661762 | 0.62 |
ENSMUST00000139341.2
ENSMUST00000017891.14 |
Ghdc
|
GH3 domain containing |
| chr19_-_37184692 | 0.62 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr11_+_118334297 | 0.62 |
ENSMUST00000133558.3
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr10_+_116137277 | 0.61 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr11_-_69586347 | 0.61 |
ENSMUST00000181261.2
|
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr7_+_78563184 | 0.60 |
ENSMUST00000121645.8
|
Isg20
|
interferon-stimulated protein |
| chr5_+_124585244 | 0.60 |
ENSMUST00000198451.2
|
Kmt5a
|
lysine methyltransferase 5A |
| chr9_-_104214920 | 0.60 |
ENSMUST00000062723.14
ENSMUST00000215852.2 |
Acpp
|
acid phosphatase, prostate |
| chr17_-_56424577 | 0.59 |
ENSMUST00000019808.12
|
Plin5
|
perilipin 5 |
| chr5_+_143166759 | 0.59 |
ENSMUST00000031574.10
|
Spdye4b
|
speedy/RINGO cell cycle regulator family, member E4B |
| chr9_+_106325860 | 0.59 |
ENSMUST00000185527.3
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr8_-_65489791 | 0.58 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr11_-_109718845 | 0.58 |
ENSMUST00000020941.11
|
1700012B07Rik
|
RIKEN cDNA 1700012B07 gene |
| chr14_+_96118660 | 0.57 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr8_-_65489834 | 0.57 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr9_+_77959206 | 0.57 |
ENSMUST00000024104.9
|
Gcm1
|
glial cells missing homolog 1 |
| chr7_+_134870237 | 0.57 |
ENSMUST00000210697.2
ENSMUST00000097983.5 |
Nps
|
neuropeptide S |
| chr11_-_20781009 | 0.56 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr2_-_30095784 | 0.56 |
ENSMUST00000113662.8
|
Kyat1
|
kynurenine aminotransferase 1 |
| chr2_-_52225146 | 0.56 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr4_-_88798438 | 0.55 |
ENSMUST00000056014.3
|
Ifne
|
interferon epsilon |
| chr1_-_65090278 | 0.54 |
ENSMUST00000161960.2
ENSMUST00000087359.6 |
Cryge
|
crystallin, gamma E |
| chr17_+_26895379 | 0.54 |
ENSMUST00000236299.2
ENSMUST00000167352.2 |
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr8_-_105981732 | 0.54 |
ENSMUST00000093217.9
ENSMUST00000161745.3 ENSMUST00000136822.3 |
B3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr8_-_95929420 | 0.53 |
ENSMUST00000212842.2
|
Kifc3
|
kinesin family member C3 |
| chr13_-_33082194 | 0.53 |
ENSMUST00000021834.11
|
Serpinb1c
|
serine (or cysteine) peptidase inhibitor, clade B, member 1c |
| chr9_+_110709301 | 0.52 |
ENSMUST00000123389.8
|
Als2cl
|
ALS2 C-terminal like |
| chr10_+_99851679 | 0.51 |
ENSMUST00000130190.8
ENSMUST00000218200.2 ENSMUST00000020129.8 |
Kitl
|
kit ligand |
| chr11_-_58213939 | 0.51 |
ENSMUST00000108829.8
|
Zfp672
|
zinc finger protein 672 |
| chr14_-_58236693 | 0.51 |
ENSMUST00000022543.10
|
Micu2
|
mitochondrial calcium uptake 2 |
| chr13_+_9143995 | 0.50 |
ENSMUST00000091829.4
|
Larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr17_+_38214413 | 0.49 |
ENSMUST00000076331.3
|
Olfr127
|
olfactory receptor 127 |
| chr3_+_27371168 | 0.49 |
ENSMUST00000046383.12
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr12_+_10419967 | 0.49 |
ENSMUST00000143739.9
ENSMUST00000002456.10 ENSMUST00000219826.2 ENSMUST00000217944.2 ENSMUST00000218339.3 ENSMUST00000118657.8 ENSMUST00000223534.2 |
Nt5c1b
|
5'-nucleotidase, cytosolic IB |
| chr13_-_34186723 | 0.48 |
ENSMUST00000167237.8
ENSMUST00000168400.8 ENSMUST00000166354.2 ENSMUST00000076532.14 ENSMUST00000171034.8 |
Serpinb6a
|
serine (or cysteine) peptidase inhibitor, clade B, member 6a |
| chr11_+_101333238 | 0.48 |
ENSMUST00000107249.8
|
Rpl27
|
ribosomal protein L27 |
| chr12_-_80690573 | 0.48 |
ENSMUST00000166931.2
ENSMUST00000218364.2 |
Erh
|
ERH mRNA splicing and mitosis factor |
| chr7_-_43885552 | 0.47 |
ENSMUST00000236952.2
|
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr9_+_106324952 | 0.47 |
ENSMUST00000215475.2
ENSMUST00000187106.7 ENSMUST00000190167.7 |
Abhd14b
|
abhydrolase domain containing 14b |
| chr5_+_114991722 | 0.47 |
ENSMUST00000031547.12
|
4930519G04Rik
|
RIKEN cDNA 4930519G04 gene |
| chr4_-_154127119 | 0.47 |
ENSMUST00000047207.7
|
Ccdc27
|
coiled-coil domain containing 27 |
| chr8_-_13888389 | 0.46 |
ENSMUST00000071308.6
|
AF366264
|
cDNA sequence AF366264 |
| chr6_+_82379768 | 0.46 |
ENSMUST00000203775.2
|
Tacr1
|
tachykinin receptor 1 |
| chr7_+_119289249 | 0.46 |
ENSMUST00000047045.10
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr2_-_168608949 | 0.46 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr2_-_168609110 | 0.46 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr7_-_98829474 | 0.46 |
ENSMUST00000207611.2
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr17_+_31515163 | 0.45 |
ENSMUST00000235972.2
ENSMUST00000165149.3 ENSMUST00000236251.2 |
Slc37a1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
| chr8_-_33374282 | 0.45 |
ENSMUST00000209107.4
ENSMUST00000209022.3 |
Nrg1
|
neuregulin 1 |
| chr2_-_28589675 | 0.45 |
ENSMUST00000124840.2
|
Spaca9
|
sperm acrosome associated 9 |
| chr2_-_28589641 | 0.45 |
ENSMUST00000102877.8
|
Spaca9
|
sperm acrosome associated 9 |
| chr17_-_56312555 | 0.45 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr11_-_109718860 | 0.44 |
ENSMUST00000106674.8
|
1700012B07Rik
|
RIKEN cDNA 1700012B07 gene |
| chrX_-_73397181 | 0.44 |
ENSMUST00000114152.2
ENSMUST00000114153.2 ENSMUST00000015433.4 |
Lage3
|
L antigen family, member 3 |
| chr14_+_20344765 | 0.44 |
ENSMUST00000223663.2
ENSMUST00000022343.6 ENSMUST00000224066.2 ENSMUST00000223941.2 ENSMUST00000224311.2 |
Nudt13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr5_-_28415361 | 0.44 |
ENSMUST00000141196.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr13_+_9143916 | 0.41 |
ENSMUST00000188211.8
ENSMUST00000188939.7 ENSMUST00000190041.7 |
Larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr17_-_71153283 | 0.41 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr16_-_58910185 | 0.41 |
ENSMUST00000215647.2
|
Olfr191
|
olfactory receptor 191 |
| chr4_+_156214969 | 0.41 |
ENSMUST00000209248.2
|
Rnf223
|
ring finger 223 |
| chr5_-_94075731 | 0.40 |
ENSMUST00000179824.2
|
Pramel36
|
PRAME like 36 |
| chr1_+_43484895 | 0.40 |
ENSMUST00000086421.9
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr15_+_25774070 | 0.39 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr6_+_82379456 | 0.39 |
ENSMUST00000032122.11
|
Tacr1
|
tachykinin receptor 1 |
| chr1_-_119924920 | 0.39 |
ENSMUST00000174370.8
ENSMUST00000174458.2 |
Cfap221
|
cilia and flagella associated protein 221 |
| chr1_+_134333720 | 0.39 |
ENSMUST00000173908.8
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr2_+_121244256 | 0.38 |
ENSMUST00000028683.14
|
Pdia3
|
protein disulfide isomerase associated 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.8 | 3.8 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.8 | 1.5 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.6 | 3.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.6 | 2.4 | GO:0032752 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.4 | 3.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.4 | 1.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.4 | 1.3 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.4 | 1.3 | GO:0060156 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.4 | 1.2 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.3 | 1.3 | GO:0060168 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.3 | 2.7 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.3 | 0.8 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.3 | 1.0 | GO:0015867 | ATP transport(GO:0015867) |
| 0.2 | 1.0 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 0.7 | GO:0046436 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 1.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 2.6 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 0.6 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.2 | 0.6 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 | 4.0 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.2 | 1.1 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 0.2 | 1.7 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.2 | 0.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 0.8 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 0.5 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.2 | 0.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.6 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.9 | GO:0046878 | angiotensin-mediated drinking behavior(GO:0003051) positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 1.5 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 2.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.6 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.4 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 1.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.7 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 0.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.4 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.1 | 0.8 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.1 | 0.5 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.7 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.5 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 0.6 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 | 1.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.4 | GO:0090096 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.3 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.2 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 0.3 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.1 | 0.5 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.1 | 1.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 5.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.9 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.0 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.1 | 1.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 5.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 0.2 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.3 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.2 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) negative regulation of locomotion involved in locomotory behavior(GO:0090327) positive regulation of eating behavior(GO:1904000) |
| 0.1 | 1.5 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 0.9 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.0 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 1.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0034309 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 2.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 4.0 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 3.0 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.7 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.4 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 1.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 1.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 2.6 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 1.4 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.3 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 4.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 1.9 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.9 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.5 | 1.6 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.4 | 1.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.4 | 3.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 1.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 2.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 3.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 1.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 1.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.9 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 4.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 1.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 3.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 1.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 5.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 1.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 8.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.9 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.5 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 4.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 1.1 | 3.2 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.6 | 1.9 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.5 | 2.4 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.4 | 1.3 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.4 | 4.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.4 | 1.2 | GO:0047312 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 0.3 | 1.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.3 | 1.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 1.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 1.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 1.7 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.2 | 1.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 0.8 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 1.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.2 | 1.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 0.7 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.2 | 0.6 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 0.4 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 1.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 2.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.5 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 4.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 2.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.0 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 2.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 1.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 1.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.5 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 3.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.7 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 9.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 3.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.2 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.1 | 0.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 1.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 4.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 1.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 2.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 2.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 4.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 2.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 8.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 6.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 3.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 5.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 4.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 1.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 4.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 3.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 4.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 3.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 3.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |