Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
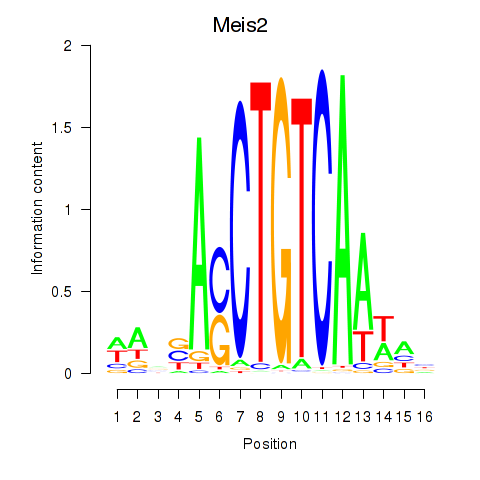
Results for Meis2
Z-value: 1.67
Transcription factors associated with Meis2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meis2
|
ENSMUSG00000027210.21 | Meis2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meis2 | mm39_v1_chr2_-_115895202_115895231 | 0.17 | 1.5e-01 | Click! |
Activity profile of Meis2 motif
Sorted Z-values of Meis2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Meis2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_119217004 | 14.67 |
ENSMUST00000047929.13
ENSMUST00000135683.3 |
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr14_-_30645503 | 14.67 |
ENSMUST00000227995.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr16_+_22877000 | 14.36 |
ENSMUST00000039492.14
ENSMUST00000023589.15 ENSMUST00000089902.8 |
Kng1
|
kininogen 1 |
| chr13_-_63036096 | 12.81 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr14_-_66246652 | 10.85 |
ENSMUST00000059970.9
|
Gulo
|
gulonolactone (L-) oxidase |
| chr5_+_137979763 | 10.29 |
ENSMUST00000035390.7
|
Azgp1
|
alpha-2-glycoprotein 1, zinc |
| chr16_-_22847808 | 10.27 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr2_+_71811526 | 10.18 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_-_4068779 | 9.83 |
ENSMUST00000003681.8
|
Sec14l2
|
SEC14-like lipid binding 2 |
| chr7_+_25872836 | 9.68 |
ENSMUST00000082214.5
|
Cyp2b9
|
cytochrome P450, family 2, subfamily b, polypeptide 9 |
| chr16_-_22847760 | 9.59 |
ENSMUST00000039338.13
|
Kng2
|
kininogen 2 |
| chr2_-_27136826 | 9.40 |
ENSMUST00000149733.8
|
Sardh
|
sarcosine dehydrogenase |
| chr11_+_83637766 | 9.39 |
ENSMUST00000070832.3
|
Wfdc21
|
WAP four-disulfide core domain 21 |
| chr16_-_22847829 | 9.23 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr6_-_85892586 | 8.98 |
ENSMUST00000174369.3
|
Nat8f1
|
N-acetyltransferase 8 (GCN5-related) family member 1 |
| chr1_+_88334678 | 8.86 |
ENSMUST00000027518.12
|
Spp2
|
secreted phosphoprotein 2 |
| chr7_-_30643444 | 8.82 |
ENSMUST00000062620.9
|
Hamp
|
hepcidin antimicrobial peptide |
| chr8_-_94063823 | 8.64 |
ENSMUST00000044602.8
|
Ces1g
|
carboxylesterase 1G |
| chr11_+_78215026 | 8.63 |
ENSMUST00000102478.4
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr7_-_126873219 | 8.48 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr12_-_103956176 | 8.38 |
ENSMUST00000151709.3
ENSMUST00000176246.3 ENSMUST00000074693.13 ENSMUST00000120251.9 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr9_-_103099262 | 8.11 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr8_+_105460627 | 8.08 |
ENSMUST00000034346.15
ENSMUST00000164182.3 |
Ces2a
|
carboxylesterase 2A |
| chr8_-_118400418 | 7.97 |
ENSMUST00000173522.8
ENSMUST00000174450.2 |
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr1_+_88139678 | 7.77 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr16_+_22710785 | 7.77 |
ENSMUST00000023583.7
ENSMUST00000232098.2 |
Ahsg
|
alpha-2-HS-glycoprotein |
| chr12_-_81015479 | 7.71 |
ENSMUST00000218162.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr5_-_87485023 | 7.62 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr16_-_22848153 | 7.49 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr9_+_106325828 | 7.22 |
ENSMUST00000217496.2
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr8_-_45747883 | 6.91 |
ENSMUST00000026907.6
|
Klkb1
|
kallikrein B, plasma 1 |
| chr16_+_22769822 | 6.78 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr19_-_43974990 | 6.77 |
ENSMUST00000026210.5
|
Cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr9_+_106325860 | 6.73 |
ENSMUST00000185527.3
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr19_-_8382424 | 6.67 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chrX_+_59044796 | 6.57 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr9_-_71075939 | 6.34 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr6_+_49799690 | 6.11 |
ENSMUST00000031843.7
|
Npy
|
neuropeptide Y |
| chr3_-_107876477 | 6.05 |
ENSMUST00000004136.10
|
Gstm3
|
glutathione S-transferase, mu 3 |
| chr6_-_106777014 | 5.98 |
ENSMUST00000013882.10
ENSMUST00000113239.10 |
Crbn
|
cereblon |
| chr8_-_118398264 | 5.88 |
ENSMUST00000037955.14
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr17_-_26309194 | 5.71 |
ENSMUST00000237265.2
ENSMUST00000236268.2 ENSMUST00000237875.2 ENSMUST00000150534.9 ENSMUST00000040907.8 |
Decr2
|
2-4-dienoyl-Coenzyme A reductase 2, peroxisomal |
| chr7_+_141503411 | 5.57 |
ENSMUST00000078200.12
ENSMUST00000018971.15 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr15_-_60793115 | 5.37 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr7_+_141503719 | 5.22 |
ENSMUST00000105989.9
ENSMUST00000075528.12 ENSMUST00000174499.8 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr3_+_10077608 | 5.21 |
ENSMUST00000029046.9
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr2_+_43445359 | 5.12 |
ENSMUST00000050511.7
|
Kynu
|
kynureninase |
| chr3_-_152687877 | 5.10 |
ENSMUST00000044278.6
|
St6galnac5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr10_+_90412827 | 5.09 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_+_97298002 | 5.09 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr17_-_33136021 | 5.03 |
ENSMUST00000054174.9
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr3_-_117153802 | 4.94 |
ENSMUST00000197743.2
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr10_+_107107477 | 4.93 |
ENSMUST00000020057.16
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr13_-_56696222 | 4.89 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr16_+_22769844 | 4.85 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr16_+_13721016 | 4.63 |
ENSMUST00000128757.8
|
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr2_+_96148418 | 4.62 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr5_-_87402659 | 4.55 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr13_+_25029088 | 4.52 |
ENSMUST00000006893.9
|
D130043K22Rik
|
RIKEN cDNA D130043K22 gene |
| chr19_-_8109346 | 4.42 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr15_+_10981833 | 4.38 |
ENSMUST00000070877.7
|
Amacr
|
alpha-methylacyl-CoA racemase |
| chr10_-_110292053 | 4.37 |
ENSMUST00000239301.2
|
Nav3
|
neuron navigator 3 |
| chr7_+_141503583 | 4.35 |
ENSMUST00000172652.8
|
Brsk2
|
BR serine/threonine kinase 2 |
| chr13_+_114954741 | 4.30 |
ENSMUST00000166104.9
ENSMUST00000166176.9 ENSMUST00000184335.8 ENSMUST00000184245.8 ENSMUST00000015680.11 ENSMUST00000184214.8 ENSMUST00000165022.9 ENSMUST00000164737.8 ENSMUST00000184781.8 ENSMUST00000183407.2 ENSMUST00000184672.8 |
Mocs2
|
molybdenum cofactor synthesis 2 |
| chr19_-_7943365 | 4.26 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr19_+_42235983 | 4.18 |
ENSMUST00000122375.8
|
Golga7b
|
golgi autoantigen, golgin subfamily a, 7B |
| chr4_+_42922253 | 4.16 |
ENSMUST00000139100.2
|
Phf24
|
PHD finger protein 24 |
| chr4_-_56990306 | 4.10 |
ENSMUST00000053681.6
|
Frrs1l
|
ferric-chelate reductase 1 like |
| chr10_-_89342493 | 4.00 |
ENSMUST00000058126.15
ENSMUST00000105296.9 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr9_-_121324744 | 3.96 |
ENSMUST00000035120.6
|
Cck
|
cholecystokinin |
| chr10_-_75633362 | 3.95 |
ENSMUST00000120177.8
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr17_-_32643067 | 3.84 |
ENSMUST00000237130.2
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr2_+_135894099 | 3.82 |
ENSMUST00000144403.8
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr15_+_16778187 | 3.79 |
ENSMUST00000026432.8
|
Cdh9
|
cadherin 9 |
| chr10_+_107107558 | 3.74 |
ENSMUST00000105280.5
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr6_+_29694181 | 3.73 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr3_-_89067462 | 3.71 |
ENSMUST00000029686.4
|
Hcn3
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
| chr7_-_86425147 | 3.71 |
ENSMUST00000001824.7
|
Folh1
|
folate hydrolase 1 |
| chr9_-_106533279 | 3.67 |
ENSMUST00000023959.13
ENSMUST00000201681.2 |
Grm2
|
glutamate receptor, metabotropic 2 |
| chr17_-_30845845 | 3.63 |
ENSMUST00000235547.2
ENSMUST00000188852.2 |
Glo1
1700097N02Rik
|
glyoxalase 1 RIKEN cDNA 1700097N02 gene |
| chr6_+_103488044 | 3.61 |
ENSMUST00000203830.3
|
Chl1
|
cell adhesion molecule L1-like |
| chr7_-_86425016 | 3.59 |
ENSMUST00000107271.10
|
Folh1
|
folate hydrolase 1 |
| chr5_-_108017019 | 3.59 |
ENSMUST00000128723.8
ENSMUST00000124034.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr14_-_70585874 | 3.59 |
ENSMUST00000152067.8
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr13_+_92562404 | 3.57 |
ENSMUST00000061594.13
|
Ankrd34b
|
ankyrin repeat domain 34B |
| chr6_+_54016543 | 3.54 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr6_-_113396271 | 3.50 |
ENSMUST00000147726.8
ENSMUST00000151618.8 ENSMUST00000060634.14 ENSMUST00000129047.8 ENSMUST00000129560.2 ENSMUST00000113092.9 |
Rpusd3
|
RNA pseudouridylate synthase domain containing 3 |
| chr1_-_121255448 | 3.49 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr1_-_121255400 | 3.48 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr8_+_107877252 | 3.46 |
ENSMUST00000034400.5
|
Cyb5b
|
cytochrome b5 type B |
| chr6_+_103488291 | 3.38 |
ENSMUST00000204321.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr7_+_143729250 | 3.35 |
ENSMUST00000105900.9
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr14_-_55950545 | 3.31 |
ENSMUST00000002389.9
ENSMUST00000226907.2 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr10_-_75633563 | 3.31 |
ENSMUST00000139724.3
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr7_+_138665900 | 3.28 |
ENSMUST00000026551.15
|
Dpysl4
|
dihydropyrimidinase-like 4 |
| chr6_+_103487973 | 3.27 |
ENSMUST00000066905.9
|
Chl1
|
cell adhesion molecule L1-like |
| chr9_-_121324705 | 3.24 |
ENSMUST00000216138.2
|
Cck
|
cholecystokinin |
| chr1_-_193052568 | 3.24 |
ENSMUST00000016323.11
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chr5_+_121798932 | 3.24 |
ENSMUST00000111765.8
|
Brap
|
BRCA1 associated protein |
| chr6_+_87405968 | 3.20 |
ENSMUST00000032125.7
|
Bmp10
|
bone morphogenetic protein 10 |
| chr1_-_184578057 | 3.19 |
ENSMUST00000068725.10
|
Mtarc2
|
mitochondrial amidoxime reducing component 2 |
| chr9_+_32027335 | 3.18 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr16_-_37681508 | 3.17 |
ENSMUST00000205931.2
|
Gm36028
|
predicted gene, 36028 |
| chr15_+_41573995 | 3.17 |
ENSMUST00000229769.2
|
Oxr1
|
oxidation resistance 1 |
| chr10_+_23770586 | 3.14 |
ENSMUST00000041416.8
|
Vnn1
|
vanin 1 |
| chr1_+_9618173 | 3.14 |
ENSMUST00000144177.8
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr5_+_24679154 | 3.13 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr17_+_35455532 | 3.11 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr15_+_7159038 | 3.06 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr2_+_27599259 | 3.06 |
ENSMUST00000100251.9
|
Rxra
|
retinoid X receptor alpha |
| chr3_-_107425316 | 3.04 |
ENSMUST00000169449.8
ENSMUST00000029499.15 |
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr7_+_91321694 | 3.00 |
ENSMUST00000238608.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr11_-_120508713 | 2.98 |
ENSMUST00000106188.4
ENSMUST00000026129.16 |
Pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr6_+_83119368 | 2.95 |
ENSMUST00000130622.8
ENSMUST00000129316.2 |
Rtkn
|
rhotekin |
| chr5_-_139115417 | 2.92 |
ENSMUST00000026973.14
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chr16_-_23709564 | 2.91 |
ENSMUST00000004480.5
|
Sst
|
somatostatin |
| chr15_-_82278223 | 2.89 |
ENSMUST00000170255.2
|
Cyp2d11
|
cytochrome P450, family 2, subfamily d, polypeptide 11 |
| chr17_+_9142941 | 2.89 |
ENSMUST00000149440.8
|
Pde10a
|
phosphodiesterase 10A |
| chr7_+_91321500 | 2.87 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr13_+_64309675 | 2.86 |
ENSMUST00000021929.10
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr19_-_4109446 | 2.85 |
ENSMUST00000189808.7
|
Gstp3
|
glutathione S-transferase pi 3 |
| chr2_-_110781268 | 2.84 |
ENSMUST00000099623.10
|
Ano3
|
anoctamin 3 |
| chr3_-_19749489 | 2.79 |
ENSMUST00000061294.5
|
Crh
|
corticotropin releasing hormone |
| chr1_+_87983099 | 2.76 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr6_+_38639945 | 2.75 |
ENSMUST00000114874.5
|
Clec2l
|
C-type lectin domain family 2, member L |
| chrX_-_149224054 | 2.75 |
ENSMUST00000059256.8
|
Tmem29
|
transmembrane protein 29 |
| chr10_-_115197775 | 2.64 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr4_+_140966810 | 2.63 |
ENSMUST00000141834.9
|
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr8_+_26091607 | 2.60 |
ENSMUST00000155861.8
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr4_-_114991478 | 2.59 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chrX_+_7588453 | 2.57 |
ENSMUST00000043045.10
ENSMUST00000207386.2 ENSMUST00000116634.9 ENSMUST00000208072.2 ENSMUST00000207589.2 ENSMUST00000208618.2 ENSMUST00000208443.2 ENSMUST00000207541.2 ENSMUST00000208528.2 ENSMUST00000115689.10 ENSMUST00000131077.9 ENSMUST00000115688.8 ENSMUST00000208156.2 |
Wdr45
Gm45208
|
WD repeat domain 45 predicted gene 45208 |
| chr3_+_81906768 | 2.56 |
ENSMUST00000107736.2
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr7_+_105203559 | 2.56 |
ENSMUST00000046983.10
|
Smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr18_+_40390013 | 2.54 |
ENSMUST00000096572.2
ENSMUST00000236889.2 |
Kctd16
|
potassium channel tetramerisation domain containing 16 |
| chr10_-_115198093 | 2.53 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr5_+_67418137 | 2.47 |
ENSMUST00000161369.3
|
Tmem33
|
transmembrane protein 33 |
| chr18_+_37223066 | 2.42 |
ENSMUST00000007584.4
|
Pcdhac1
|
protocadherin alpha subfamily C, 1 |
| chr2_+_30156523 | 2.41 |
ENSMUST00000091132.13
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr2_-_30095805 | 2.40 |
ENSMUST00000113663.9
ENSMUST00000044038.10 |
Kyat1
|
kynurenine aminotransferase 1 |
| chr4_+_100336003 | 2.38 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr1_-_107206091 | 2.37 |
ENSMUST00000166100.2
ENSMUST00000027565.5 |
Serpinb3b
Serpinb3c
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3B serine (or cysteine) peptidase inhibitor, clade B, member 3C |
| chr2_+_132688558 | 2.33 |
ENSMUST00000028835.13
ENSMUST00000110122.10 |
Crls1
|
cardiolipin synthase 1 |
| chr5_-_122917341 | 2.30 |
ENSMUST00000198257.5
ENSMUST00000199599.2 ENSMUST00000196742.2 ENSMUST00000200109.5 ENSMUST00000111668.8 |
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr3_+_107137924 | 2.29 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr3_-_110051253 | 2.27 |
ENSMUST00000133268.9
ENSMUST00000051253.4 |
Ntng1
|
netrin G1 |
| chr7_+_119773070 | 2.26 |
ENSMUST00000033201.7
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr7_+_3352159 | 2.26 |
ENSMUST00000172109.4
|
Prkcg
|
protein kinase C, gamma |
| chr4_-_129472328 | 2.25 |
ENSMUST00000052835.9
|
Fam167b
|
family with sequence similarity 167, member B |
| chr7_-_132178101 | 2.24 |
ENSMUST00000084500.8
|
Oat
|
ornithine aminotransferase |
| chr10_-_29411857 | 2.23 |
ENSMUST00000092623.5
|
Rspo3
|
R-spondin 3 |
| chrX_+_136552469 | 2.21 |
ENSMUST00000075471.4
|
Il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr2_-_30095784 | 2.21 |
ENSMUST00000113662.8
|
Kyat1
|
kynurenine aminotransferase 1 |
| chr11_-_83959999 | 2.20 |
ENSMUST00000138208.2
|
Dusp14
|
dual specificity phosphatase 14 |
| chr11_-_46203047 | 2.19 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chrX_+_7588505 | 2.19 |
ENSMUST00000207675.2
ENSMUST00000116633.9 ENSMUST00000208996.2 ENSMUST00000144148.4 ENSMUST00000125991.9 ENSMUST00000148624.8 |
Wdr45
|
WD repeat domain 45 |
| chr8_-_70686746 | 2.18 |
ENSMUST00000130319.2
|
Armc6
|
armadillo repeat containing 6 |
| chr1_-_24626492 | 2.17 |
ENSMUST00000051344.6
ENSMUST00000115244.9 |
Col19a1
|
collagen, type XIX, alpha 1 |
| chr1_-_121255503 | 2.16 |
ENSMUST00000160688.2
|
Insig2
|
insulin induced gene 2 |
| chrX_+_141011695 | 2.12 |
ENSMUST00000112913.2
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr3_-_92697821 | 2.11 |
ENSMUST00000186525.2
|
Lce1j
|
late cornified envelope 1J |
| chr13_+_24511387 | 2.10 |
ENSMUST00000224953.2
ENSMUST00000050859.13 ENSMUST00000167746.8 ENSMUST00000224819.2 |
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr8_-_3767547 | 2.08 |
ENSMUST00000058040.7
|
Clec4g
|
C-type lectin domain family 4, member g |
| chr3_+_85481416 | 2.07 |
ENSMUST00000107672.8
ENSMUST00000127348.8 ENSMUST00000107674.2 |
Gatb
|
glutamyl-tRNA(Gln) amidotransferase, subunit B |
| chr7_+_3352019 | 2.06 |
ENSMUST00000100301.11
|
Prkcg
|
protein kinase C, gamma |
| chr1_-_184615415 | 2.03 |
ENSMUST00000048308.6
|
C130074G19Rik
|
RIKEN cDNA C130074G19 gene |
| chr1_-_105284407 | 2.03 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr4_+_42091207 | 2.01 |
ENSMUST00000178882.2
|
Gm3893
|
predicted gene 3893 |
| chr1_-_180023518 | 2.00 |
ENSMUST00000162769.8
ENSMUST00000161379.2 ENSMUST00000027766.13 ENSMUST00000161814.8 |
Coq8a
|
coenzyme Q8A |
| chr8_-_68363564 | 2.00 |
ENSMUST00000093468.12
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr8_+_71261073 | 1.97 |
ENSMUST00000000808.8
ENSMUST00000212657.2 ENSMUST00000212146.2 |
Il12rb1
|
interleukin 12 receptor, beta 1 |
| chr11_-_83469446 | 1.97 |
ENSMUST00000019266.6
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr6_-_36787096 | 1.94 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr3_-_53771185 | 1.94 |
ENSMUST00000122330.2
ENSMUST00000146598.8 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr18_+_56565188 | 1.91 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr5_+_105879914 | 1.91 |
ENSMUST00000154807.2
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr4_-_129334593 | 1.88 |
ENSMUST00000053042.6
ENSMUST00000106046.8 |
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr15_+_4756684 | 1.86 |
ENSMUST00000161997.8
ENSMUST00000022788.15 |
C6
|
complement component 6 |
| chr9_-_107419309 | 1.84 |
ENSMUST00000195235.6
|
Cyb561d2
|
cytochrome b-561 domain containing 2 |
| chr5_-_100307130 | 1.84 |
ENSMUST00000139520.3
|
Tmem150c
|
transmembrane protein 150C |
| chr11_+_77409392 | 1.82 |
ENSMUST00000147386.2
|
Abhd15
|
abhydrolase domain containing 15 |
| chr14_-_63781381 | 1.82 |
ENSMUST00000058679.7
|
Mtmr9
|
myotubularin related protein 9 |
| chr8_-_68270936 | 1.81 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr7_+_6401706 | 1.81 |
ENSMUST00000161855.8
|
Smim17
|
small integral membrane protein 17 |
| chr1_+_156443472 | 1.81 |
ENSMUST00000190749.2
|
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr8_+_105573693 | 1.79 |
ENSMUST00000055052.6
|
Ces2c
|
carboxylesterase 2C |
| chr18_+_63841756 | 1.79 |
ENSMUST00000072726.7
ENSMUST00000235648.2 ENSMUST00000236879.2 |
Wdr7
|
WD repeat domain 7 |
| chr3_-_84178089 | 1.78 |
ENSMUST00000054990.11
|
Trim2
|
tripartite motif-containing 2 |
| chr11_+_55095144 | 1.75 |
ENSMUST00000108872.9
ENSMUST00000147506.8 ENSMUST00000020499.14 |
Slc36a1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr9_-_106124917 | 1.72 |
ENSMUST00000112524.9
ENSMUST00000219129.2 |
Alas1
|
aminolevulinic acid synthase 1 |
| chr3_+_134534754 | 1.69 |
ENSMUST00000029822.6
|
Tacr3
|
tachykinin receptor 3 |
| chr8_-_72178340 | 1.69 |
ENSMUST00000153800.8
ENSMUST00000146100.8 |
Fcho1
|
FCH domain only 1 |
| chr7_+_143376871 | 1.68 |
ENSMUST00000128454.8
ENSMUST00000073878.12 |
Dhcr7
|
7-dehydrocholesterol reductase |
| chr9_-_66882669 | 1.67 |
ENSMUST00000215172.2
ENSMUST00000034929.7 |
Lactb
|
lactamase, beta |
| chr10_-_40948275 | 1.67 |
ENSMUST00000061796.8
|
Gpr6
|
G protein-coupled receptor 6 |
| chr7_+_44499818 | 1.67 |
ENSMUST00000136232.2
ENSMUST00000207223.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr5_-_36987917 | 1.66 |
ENSMUST00000031002.10
|
Man2b2
|
mannosidase 2, alpha B2 |
| chr2_+_120807498 | 1.66 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr6_-_83654789 | 1.64 |
ENSMUST00000037882.8
|
Cd207
|
CD207 antigen |
| chr14_+_15369152 | 1.64 |
ENSMUST00000167923.8
|
Gm3696
|
predicted gene 3696 |
| chr1_+_87983189 | 1.62 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0097037 | heme export(GO:0097037) |
| 3.2 | 12.8 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 3.1 | 9.4 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 3.0 | 9.0 | GO:0018003 | peptidyl-lysine N6-acetylation(GO:0018003) |
| 2.9 | 8.6 | GO:0034378 | chylomicron assembly(GO:0034378) regulation of chylomicron remnant clearance(GO:0090320) |
| 2.6 | 7.8 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 2.4 | 7.3 | GO:0018900 | dichloromethane metabolic process(GO:0018900) |
| 2.2 | 8.8 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 2.2 | 10.8 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 2.0 | 10.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 1.7 | 5.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 1.6 | 6.3 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.5 | 4.6 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 1.4 | 8.7 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 1.4 | 6.9 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.2 | 8.5 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 1.2 | 3.6 | GO:0030070 | insulin processing(GO:0030070) |
| 1.0 | 15.4 | GO:0015747 | urate transport(GO:0015747) |
| 1.0 | 8.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 1.0 | 4.0 | GO:0038183 | histone H3-R17 methylation(GO:0034971) bile acid signaling pathway(GO:0038183) |
| 0.9 | 2.8 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.9 | 12.0 | GO:0006063 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) cellular glucuronidation(GO:0052695) |
| 0.9 | 3.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.8 | 51.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.8 | 2.5 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.8 | 4.6 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.8 | 2.3 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.7 | 4.3 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.7 | 2.7 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.6 | 3.8 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.6 | 1.9 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.6 | 3.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.6 | 3.6 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.6 | 1.7 | GO:0016131 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.5 | 8.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.5 | 4.3 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.5 | 2.1 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.5 | 5.7 | GO:0015824 | proline transport(GO:0015824) |
| 0.5 | 9.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.5 | 1.5 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.5 | 9.8 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.5 | 1.9 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.5 | 0.9 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.5 | 5.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.4 | 3.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.4 | 13.5 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.4 | 2.0 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.4 | 1.9 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.4 | 17.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.4 | 1.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.4 | 1.5 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.4 | 3.6 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.4 | 5.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.4 | 3.6 | GO:0010612 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.4 | 3.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.3 | 4.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 4.4 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.3 | 3.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.3 | 1.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.3 | 9.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 1.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 1.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 3.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.3 | 1.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.3 | 1.4 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.3 | 8.4 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.3 | 15.4 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.3 | 5.9 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 1.6 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.3 | 1.6 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.2 | 0.7 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.2 | 2.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.2 | 7.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 0.7 | GO:2000769 | regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.2 | 4.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 1.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.2 | 6.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 9.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 1.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 2.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 5.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 0.6 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.2 | 2.5 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 | 8.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 3.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 1.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.7 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 3.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 1.4 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 3.1 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 4.8 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.2 | 0.9 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.2 | 3.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 14.2 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.2 | 0.5 | GO:1900864 | tRNA wobble position uridine thiolation(GO:0002143) mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 1.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 4.6 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.2 | 1.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.2 | 1.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.6 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 1.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 1.1 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 | 6.2 | GO:0006760 | folic acid-containing compound metabolic process(GO:0006760) |
| 0.1 | 2.8 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.1 | 5.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.0 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 3.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 13.9 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 2.6 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 2.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.8 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.1 | 1.3 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 0.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) regulation of intracellular mRNA localization(GO:1904580) |
| 0.1 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 3.0 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 0.4 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.1 | 4.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 6.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 6.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.3 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.9 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 1.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.3 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.1 | 4.1 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 6.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.3 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 4.5 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.4 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.7 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.3 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 1.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.9 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.9 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 3.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.4 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 3.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.7 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 6.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.5 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 13.9 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.1 | 1.0 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 2.9 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.1 | 0.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 2.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 2.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.9 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 1.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 1.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.2 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 3.6 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 4.9 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 0.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 2.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 3.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.2 | GO:2000630 | hair follicle cell proliferation(GO:0071335) positive regulation of miRNA metabolic process(GO:2000630) |
| 0.1 | 1.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.5 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.2 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.6 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.4 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 3.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 1.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 4.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.8 | GO:0044783 | mitotic G1 DNA damage checkpoint(GO:0031571) G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.0 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 3.8 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0070893 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.3 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 2.3 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 | 0.5 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.7 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 1.4 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 1.1 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 3.6 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 2.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 2.8 | 13.9 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.4 | 4.3 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 1.3 | 12.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 1.1 | 9.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.1 | 8.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.7 | 8.1 | GO:0097433 | dense body(GO:0097433) |
| 0.6 | 1.9 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.6 | 4.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.5 | 7.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 6.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.4 | 7.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.4 | 3.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 1.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.3 | 4.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.3 | 3.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 4.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 4.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 69.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 0.7 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 1.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 3.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 2.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 1.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 0.8 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.2 | 1.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.8 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 5.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 1.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 1.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 0.9 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 2.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.7 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.6 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 1.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 5.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 5.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 16.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 1.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 8.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 0.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 25.7 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 1.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 11.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0035859 | Seh1-associated complex(GO:0035859) |
| 0.0 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 56.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 2.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 6.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.6 | GO:0022624 | proteasome regulatory particle(GO:0005838) proteasome accessory complex(GO:0022624) |
| 0.0 | 4.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.7 | GO:0031968 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 5.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 10.6 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 6.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.9 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.4 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 2.8 | 8.5 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 2.8 | 11.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 2.4 | 7.3 | GO:0047651 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 1.8 | 14.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 1.7 | 5.0 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 1.5 | 4.6 | GO:0047312 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 1.4 | 4.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.4 | 5.7 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.3 | 7.8 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 1.3 | 5.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.3 | 6.3 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 1.2 | 8.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.2 | 8.1 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 1.0 | 3.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.0 | 7.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.9 | 15.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.9 | 2.7 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.9 | 13.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.9 | 8.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.8 | 5.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.8 | 9.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.8 | 12.8 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.8 | 4.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.8 | 15.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.7 | 2.1 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.7 | 2.8 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.7 | 3.9 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.6 | 3.8 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.6 | 2.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.6 | 1.9 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.6 | 3.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.6 | 24.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 2.4 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.6 | 5.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.6 | 65.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.6 | 1.7 | GO:0009918 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.6 | 2.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.5 | 3.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.5 | 1.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.5 | 2.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.4 | 15.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 1.7 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.4 | 2.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 13.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 2.0 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.4 | 3.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.4 | 7.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.4 | 1.5 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.4 | 13.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.4 | 3.6 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.3 | 3.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 4.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.3 | 3.0 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.3 | 4.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.3 | 1.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.3 | 1.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 3.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 3.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 3.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.3 | 1.3 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 2.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 1.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 0.7 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 0.2 | 3.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 3.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 4.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 2.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 3.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 2.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 1.2 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 4.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.2 | 14.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 2.9 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.8 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 2.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 4.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 5.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 3.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 3.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 8.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 20.8 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.1 | 0.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 5.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 3.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.4 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 1.1 | GO:0061135 | endopeptidase regulator activity(GO:0061135) |
| 0.1 | 3.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 3.0 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.4 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 1.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 1.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 3.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 5.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 2.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 14.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 1.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 3.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 2.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 1.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 5.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.4 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 2.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 4.9 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 6.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 1.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 2.4 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 9.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.4 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 3.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 1.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 16.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 40.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 9.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 7.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 4.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 6.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 2.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 4.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 3.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 27.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.7 | 7.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.7 | 7.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.5 | 6.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 22.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 5.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 4.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 10.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 4.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 14.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 3.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 3.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 5.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 3.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 11.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 18.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 3.6 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 1.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 4.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 2.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 3.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 5.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |