Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Meox1
Z-value: 0.70
Transcription factors associated with Meox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox1
|
ENSMUSG00000001493.10 | Meox1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meox1 | mm39_v1_chr11_-_101785181_101785200 | 0.07 | 5.7e-01 | Click! |
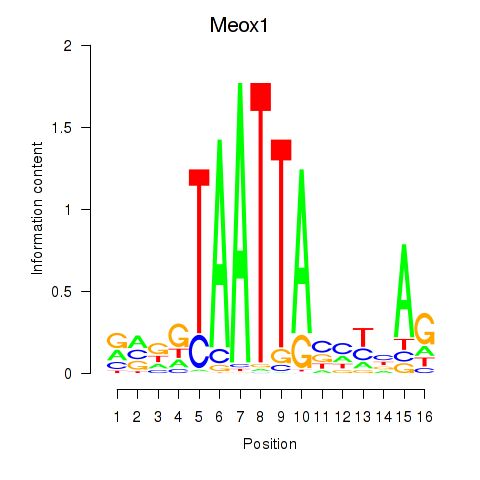
Activity profile of Meox1 motif
Sorted Z-values of Meox1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Meox1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_106126794 | 9.21 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr2_-_58050494 | 5.09 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr7_-_25112256 | 4.32 |
ENSMUST00000200880.4
ENSMUST00000074040.4 |
Cxcl17
|
chemokine (C-X-C motif) ligand 17 |
| chr3_-_15902583 | 3.83 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr11_+_115802828 | 3.75 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr8_-_62576140 | 3.72 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr8_-_25314032 | 3.62 |
ENSMUST00000050300.15
ENSMUST00000209935.2 |
Adam5
|
a disintegrin and metallopeptidase domain 5 |
| chr10_+_115653152 | 3.29 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr10_+_127734384 | 2.51 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chrX_+_149330371 | 2.50 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr12_-_55061117 | 2.44 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr19_+_34078333 | 2.43 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr2_+_174292471 | 2.37 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr3_+_92586546 | 2.35 |
ENSMUST00000047055.4
|
Lce1c
|
late cornified envelope 1C |
| chr1_-_138103021 | 2.31 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr2_-_110193502 | 2.29 |
ENSMUST00000099626.5
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr2_-_84255602 | 2.23 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr11_-_69786324 | 2.22 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr1_-_138102972 | 2.12 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr12_+_113038376 | 2.11 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22 |
| chr3_-_15640045 | 2.02 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr8_-_72763462 | 2.01 |
ENSMUST00000003574.5
|
Cyp4f18
|
cytochrome P450, family 4, subfamily f, polypeptide 18 |
| chr5_-_134776101 | 2.00 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chrX_+_47235313 | 1.99 |
ENSMUST00000033427.7
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr10_-_85847697 | 1.86 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr6_-_124756645 | 1.85 |
ENSMUST00000147669.2
ENSMUST00000128697.8 ENSMUST00000032218.10 ENSMUST00000112475.9 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr15_+_37233280 | 1.83 |
ENSMUST00000161405.8
ENSMUST00000022895.15 ENSMUST00000161532.2 |
Grhl2
|
grainyhead like transcription factor 2 |
| chr18_-_10182007 | 1.75 |
ENSMUST00000067947.7
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr18_-_7273578 | 1.72 |
ENSMUST00000234281.2
ENSMUST00000234196.2 |
Odad2
|
outer dynein arm docking complex subunit 2 |
| chr4_+_111863441 | 1.72 |
ENSMUST00000162885.8
ENSMUST00000117379.9 ENSMUST00000161389.8 ENSMUST00000162158.2 |
Skint1
|
selection and upkeep of intraepithelial T cells 1 |
| chr2_+_84818538 | 1.70 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr17_+_88748139 | 1.69 |
ENSMUST00000112238.9
ENSMUST00000155640.2 |
Foxn2
|
forkhead box N2 |
| chr13_+_34923589 | 1.68 |
ENSMUST00000221037.2
|
Fam50b
|
family with sequence similarity 50, member B |
| chr4_+_102843540 | 1.67 |
ENSMUST00000030248.12
ENSMUST00000125417.9 ENSMUST00000169211.3 |
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr11_+_88964667 | 1.64 |
ENSMUST00000100619.11
|
Gm525
|
predicted gene 525 |
| chr7_+_45271229 | 1.62 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr15_-_101801351 | 1.54 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr14_+_26414422 | 1.54 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr10_+_87041814 | 1.54 |
ENSMUST00000189775.2
|
1700113H08Rik
|
RIKEN cDNA 1700113H08 gene |
| chr6_-_73446560 | 1.52 |
ENSMUST00000070163.6
|
4931417E11Rik
|
RIKEN cDNA 4931417E11 gene |
| chr5_-_62923463 | 1.48 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr1_-_144427302 | 1.44 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr18_+_46874920 | 1.41 |
ENSMUST00000025357.9
ENSMUST00000225520.2 |
Ap3s1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr3_-_15491482 | 1.39 |
ENSMUST00000099201.9
ENSMUST00000194144.3 ENSMUST00000192700.3 |
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr7_-_43309563 | 1.31 |
ENSMUST00000032667.10
|
Siglece
|
sialic acid binding Ig-like lectin E |
| chr5_+_20112771 | 1.29 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_-_34294377 | 1.23 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr2_+_110551976 | 1.20 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr14_+_96118660 | 1.19 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr9_-_58648826 | 1.18 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr2_-_119985078 | 1.18 |
ENSMUST00000028755.8
|
Ehd4
|
EH-domain containing 4 |
| chr18_+_52748978 | 1.15 |
ENSMUST00000072666.4
ENSMUST00000209270.2 |
Zfp474
|
zinc finger protein 474 |
| chr4_-_14621805 | 1.15 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr2_+_110551927 | 1.14 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr7_-_24423715 | 1.14 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr10_-_62363217 | 1.13 |
ENSMUST00000160987.8
|
Srgn
|
serglycin |
| chr15_-_101651534 | 1.12 |
ENSMUST00000023710.6
|
Krt71
|
keratin 71 |
| chr3_-_130524024 | 1.12 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr14_-_122153185 | 1.09 |
ENSMUST00000055475.9
|
Gpr18
|
G protein-coupled receptor 18 |
| chr2_+_110551685 | 1.09 |
ENSMUST00000111016.9
|
Muc15
|
mucin 15 |
| chr3_-_54962922 | 1.08 |
ENSMUST00000197238.5
|
Ccna1
|
cyclin A1 |
| chr10_-_62363192 | 1.08 |
ENSMUST00000160643.8
|
Srgn
|
serglycin |
| chr6_-_56546088 | 1.07 |
ENSMUST00000203372.3
|
Pde1c
|
phosphodiesterase 1C |
| chr9_-_71803354 | 1.07 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr19_-_5610628 | 1.07 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr10_-_103072141 | 1.05 |
ENSMUST00000123364.2
ENSMUST00000166240.8 ENSMUST00000020043.12 |
Lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr10_+_57527084 | 1.04 |
ENSMUST00000175852.2
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr14_+_54119038 | 1.04 |
ENSMUST00000103676.9
ENSMUST00000187163.2 |
Trdv1
|
T cell receptor delta variable 1 |
| chr2_-_27365633 | 1.03 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr2_-_28806639 | 1.03 |
ENSMUST00000113847.3
|
Barhl1
|
BarH like homeobox 1 |
| chr12_-_104439589 | 1.01 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr3_-_54962899 | 0.99 |
ENSMUST00000199144.5
|
Ccna1
|
cyclin A1 |
| chr18_+_46874970 | 0.96 |
ENSMUST00000224622.2
|
Ap3s1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr1_-_4479445 | 0.95 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr13_-_32967937 | 0.95 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr5_-_65855511 | 0.93 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr8_-_87307294 | 0.93 |
ENSMUST00000131423.8
ENSMUST00000152438.2 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr10_+_57527073 | 0.93 |
ENSMUST00000066028.13
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr14_+_64229914 | 0.93 |
ENSMUST00000058229.6
|
Rp1l1
|
retinitis pigmentosa 1 homolog like 1 |
| chr3_-_130523954 | 0.91 |
ENSMUST00000196202.5
ENSMUST00000133802.6 ENSMUST00000062601.14 ENSMUST00000200517.2 |
Rpl34
|
ribosomal protein L34 |
| chr4_-_14621669 | 0.91 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr10_+_32959472 | 0.91 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr7_+_130936227 | 0.90 |
ENSMUST00000207784.2
|
Fam24a
|
family with sequence similarity 24, member A |
| chr17_-_45970238 | 0.89 |
ENSMUST00000120717.8
|
Capn11
|
calpain 11 |
| chr7_-_3901119 | 0.89 |
ENSMUST00000070639.8
|
Gm14548
|
predicted gene 14548 |
| chr17_+_34258411 | 0.88 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr7_+_130936306 | 0.87 |
ENSMUST00000084505.5
|
Fam24a
|
family with sequence similarity 24, member A |
| chr1_+_92500847 | 0.87 |
ENSMUST00000074859.3
|
Olfr1413
|
olfactory receptor 1413 |
| chr10_+_41179966 | 0.87 |
ENSMUST00000173494.4
|
Ak9
|
adenylate kinase 9 |
| chr13_-_55169000 | 0.84 |
ENSMUST00000153665.8
|
Hk3
|
hexokinase 3 |
| chr14_+_32507920 | 0.83 |
ENSMUST00000039191.8
ENSMUST00000227060.2 ENSMUST00000228481.2 |
Tmem273
|
transmembrane protein 273 |
| chr5_-_23881353 | 0.82 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr16_+_33071784 | 0.81 |
ENSMUST00000023502.6
|
Snx4
|
sorting nexin 4 |
| chr7_+_19102423 | 0.78 |
ENSMUST00000132655.2
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr1_-_4430481 | 0.78 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr8_+_41246310 | 0.78 |
ENSMUST00000056331.8
|
Adam20
|
a disintegrin and metallopeptidase domain 20 |
| chr5_-_86893645 | 0.77 |
ENSMUST00000161306.2
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr11_+_116734104 | 0.76 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr3_-_16060545 | 0.76 |
ENSMUST00000194367.6
|
Gm5150
|
predicted gene 5150 |
| chr11_-_114851243 | 0.76 |
ENSMUST00000092466.13
ENSMUST00000061637.4 |
Cd300c
|
CD300C molecule |
| chr5_-_123127346 | 0.76 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr1_+_82817794 | 0.73 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr2_+_164455438 | 0.71 |
ENSMUST00000094346.3
|
Wfdc6b
|
WAP four-disulfide core domain 6B |
| chr1_-_69726384 | 0.71 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr6_+_8520006 | 0.71 |
ENSMUST00000162567.8
ENSMUST00000161217.8 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr9_-_36637923 | 0.71 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr7_+_24776630 | 0.71 |
ENSMUST00000179556.2
ENSMUST00000053410.11 |
Zfp574
|
zinc finger protein 574 |
| chr4_-_41464816 | 0.69 |
ENSMUST00000108055.9
ENSMUST00000154535.8 ENSMUST00000030148.6 |
Kif24
|
kinesin family member 24 |
| chr12_+_108572015 | 0.69 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr9_-_36637670 | 0.68 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr1_-_20890437 | 0.68 |
ENSMUST00000053266.11
|
Mcm3
|
minichromosome maintenance complex component 3 |
| chr13_-_19521337 | 0.67 |
ENSMUST00000103563.3
|
Trgv2
|
T cell receptor gamma variable 2 |
| chr9_+_94551929 | 0.65 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr9_-_46231282 | 0.65 |
ENSMUST00000159565.8
|
4931429L15Rik
|
RIKEN cDNA 4931429L15 gene |
| chr4_-_14621497 | 0.64 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr7_-_12829100 | 0.64 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr16_-_58620631 | 0.64 |
ENSMUST00000206205.3
|
Olfr173
|
olfactory receptor 173 |
| chr10_-_129738595 | 0.64 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr3_-_16060491 | 0.63 |
ENSMUST00000108347.3
|
Gm5150
|
predicted gene 5150 |
| chr8_+_23901506 | 0.63 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr1_+_171386752 | 0.62 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr9_-_70842090 | 0.62 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr5_-_137015683 | 0.61 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr9_-_103357564 | 0.60 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr12_+_105996961 | 0.59 |
ENSMUST00000220629.2
|
Vrk1
|
vaccinia related kinase 1 |
| chr16_+_34510918 | 0.58 |
ENSMUST00000023532.7
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr4_-_117539431 | 0.57 |
ENSMUST00000102687.4
|
Dmap1
|
DNA methyltransferase 1-associated protein 1 |
| chr14_+_54082691 | 0.57 |
ENSMUST00000103674.6
|
Trav19
|
T cell receptor alpha variable 19 |
| chr9_+_51027300 | 0.56 |
ENSMUST00000114431.4
ENSMUST00000213117.2 |
Btg4
|
BTG anti-proliferation factor 4 |
| chr7_-_119058489 | 0.54 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr6_+_103674695 | 0.52 |
ENSMUST00000205098.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr18_-_46874611 | 0.52 |
ENSMUST00000035648.6
|
Atg12
|
autophagy related 12 |
| chr4_+_62443606 | 0.51 |
ENSMUST00000062145.2
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr1_-_134883645 | 0.51 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr11_-_99519057 | 0.51 |
ENSMUST00000081007.7
|
Krtap4-1
|
keratin associated protein 4-1 |
| chr4_+_8690398 | 0.50 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr14_+_62529924 | 0.50 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr16_+_34815177 | 0.49 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase |
| chr12_-_79343040 | 0.49 |
ENSMUST00000218377.2
ENSMUST00000021547.8 |
Zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr15_-_63869818 | 0.49 |
ENSMUST00000164532.3
|
Cyrib
|
CYFIP related Rac1 interactor B |
| chr1_+_150195158 | 0.48 |
ENSMUST00000165062.8
ENSMUST00000191228.7 ENSMUST00000186572.7 ENSMUST00000185698.2 |
Pdc
|
phosducin |
| chr16_-_48592319 | 0.48 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr16_+_34511073 | 0.47 |
ENSMUST00000231609.2
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr6_+_54794433 | 0.47 |
ENSMUST00000127331.2
|
Znrf2
|
zinc and ring finger 2 |
| chr18_+_12874368 | 0.46 |
ENSMUST00000235000.2
ENSMUST00000115857.9 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr13_-_95661726 | 0.45 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr3_-_92646403 | 0.45 |
ENSMUST00000192027.2
|
Gm38119
|
predicted gene, 38119 |
| chr9_+_40092216 | 0.45 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr14_+_54198389 | 0.43 |
ENSMUST00000103678.4
|
Trdv2-2
|
T cell receptor delta variable 2-2 |
| chr2_-_164455520 | 0.43 |
ENSMUST00000094351.11
ENSMUST00000109338.2 |
Wfdc8
|
WAP four-disulfide core domain 8 |
| chr13_+_21900554 | 0.40 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13 |
| chr9_+_123195986 | 0.39 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr1_-_126665904 | 0.38 |
ENSMUST00000160693.2
|
Nckap5
|
NCK-associated protein 5 |
| chr5_+_107656810 | 0.37 |
ENSMUST00000160160.6
|
Gm42669
|
predicted gene 42669 |
| chr11_-_114892706 | 0.36 |
ENSMUST00000092464.10
|
Cd300c2
|
CD300C molecule 2 |
| chr9_-_18667005 | 0.36 |
ENSMUST00000066997.4
|
Olfr24
|
olfactory receptor 24 |
| chr10_-_75946790 | 0.36 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr2_-_164585102 | 0.35 |
ENSMUST00000103096.10
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr14_-_7766684 | 0.35 |
ENSMUST00000225630.3
ENSMUST00000225979.3 ENSMUST00000226079.3 ENSMUST00000223607.3 ENSMUST00000223761.3 ENSMUST00000223981.3 ENSMUST00000224222.3 ENSMUST00000225175.3 ENSMUST00000225238.3 ENSMUST00000225232.3 |
Slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr5_+_20112500 | 0.34 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_+_132832401 | 0.34 |
ENSMUST00000032315.3
|
Tas2r116
|
taste receptor, type 2, member 116 |
| chrX_+_135828069 | 0.33 |
ENSMUST00000059808.5
|
H2bw2
|
H2B.W histone 2 |
| chr10_+_111342147 | 0.32 |
ENSMUST00000164773.2
|
Phlda1
|
pleckstrin homology like domain, family A, member 1 |
| chr8_+_22682816 | 0.31 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chrX_+_100473161 | 0.29 |
ENSMUST00000033673.7
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chr1_+_78286946 | 0.29 |
ENSMUST00000036172.10
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr7_-_140590605 | 0.28 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr18_-_36877571 | 0.28 |
ENSMUST00000014438.5
|
Ndufa2
|
NADH:ubiquinone oxidoreductase subunit A2 |
| chr2_-_45007407 | 0.28 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr17_-_50600620 | 0.27 |
ENSMUST00000010736.9
|
Dazl
|
deleted in azoospermia-like |
| chr7_+_29931309 | 0.26 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr6_-_41752111 | 0.26 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr3_+_102927901 | 0.26 |
ENSMUST00000198180.5
ENSMUST00000197827.5 ENSMUST00000199240.5 ENSMUST00000199420.5 ENSMUST00000199571.5 ENSMUST00000197488.5 |
Csde1
|
cold shock domain containing E1, RNA binding |
| chr3_-_129834788 | 0.25 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr1_-_75208734 | 0.25 |
ENSMUST00000180101.3
|
A630095N17Rik
|
RIKEN cDNA A630095N17 gene |
| chr6_+_72074545 | 0.25 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr16_+_49620883 | 0.25 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr10_-_81186025 | 0.24 |
ENSMUST00000122993.8
|
Hmg20b
|
high mobility group 20B |
| chr10_-_129710624 | 0.24 |
ENSMUST00000081367.2
|
Olfr814
|
olfactory receptor 814 |
| chr18_-_67378512 | 0.24 |
ENSMUST00000237631.2
|
Mppe1
|
metallophosphoesterase 1 |
| chr4_-_118549953 | 0.24 |
ENSMUST00000216226.2
|
Olfr1342
|
olfactory receptor 1342 |
| chrX_-_91643178 | 0.22 |
ENSMUST00000113955.2
|
Mageb18
|
MAGE family member B18 |
| chr2_+_89642395 | 0.22 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr2_-_86791037 | 0.22 |
ENSMUST00000213456.2
|
Olfr1099
|
olfactory receptor 1099 |
| chr10_-_81186222 | 0.22 |
ENSMUST00000020454.11
ENSMUST00000105324.9 ENSMUST00000154609.3 ENSMUST00000105323.8 |
Hmg20b
|
high mobility group 20B |
| chr18_+_12874390 | 0.20 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr5_-_96312006 | 0.20 |
ENSMUST00000137207.2
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr12_-_99529767 | 0.20 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr5_+_20112704 | 0.20 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr8_+_84262409 | 0.19 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr5_+_138185747 | 0.19 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr6_-_69162381 | 0.19 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr10_-_81186137 | 0.19 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr16_-_63684477 | 0.19 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr2_+_155593030 | 0.18 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chr3_-_144514386 | 0.17 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr7_-_103094646 | 0.17 |
ENSMUST00000215417.2
|
Olfr605
|
olfactory receptor 605 |
| chr7_+_29931735 | 0.16 |
ENSMUST00000108193.2
ENSMUST00000108192.2 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr10_+_81554753 | 0.15 |
ENSMUST00000085664.6
|
Zfp433
|
zinc finger protein 433 |
| chr10_+_97400990 | 0.15 |
ENSMUST00000038160.6
|
Lum
|
lumican |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 1.0 | 9.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.7 | 2.0 | GO:1901662 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.6 | 1.8 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.6 | 2.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.6 | 2.2 | GO:0033377 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.5 | 1.4 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.4 | 1.7 | GO:0045575 | basophil activation(GO:0045575) |
| 0.4 | 1.2 | GO:0018931 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.4 | 1.1 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.4 | 1.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.3 | 1.3 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.3 | 2.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 1.8 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 0.9 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.2 | 0.6 | GO:2000040 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 | 0.8 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 4.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.2 | 0.5 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.2 | 1.8 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 0.5 | GO:1902566 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.2 | 0.9 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.6 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.1 | 2.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.4 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.5 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.6 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.1 | 1.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 1.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 2.0 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 2.4 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 2.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.8 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 1.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.6 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.6 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.5 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 1.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.1 | GO:1900239 | regulation of phenotypic switching(GO:1900239) |
| 0.1 | 1.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.8 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 0.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 3.5 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 1.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 2.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 1.2 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.8 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 1.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.9 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.4 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 3.1 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 1.4 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 2.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.9 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.3 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.8 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 1.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.3 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.3 | GO:0046597 | response to interferon-alpha(GO:0035455) negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 4.6 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.8 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.5 | 2.1 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.4 | 2.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 3.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 0.5 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 0.6 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.1 | 2.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 1.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 2.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.6 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 3.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 5.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.8 | 2.5 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.7 | 2.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.7 | 2.0 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.3 | 1.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 1.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.9 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.2 | 0.8 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 0.6 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.2 | 0.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 1.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 5.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.4 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 1.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 2.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.8 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 4.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.1 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.2 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 2.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 3.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 3.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 2.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 4.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 7.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 1.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 2.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |