Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
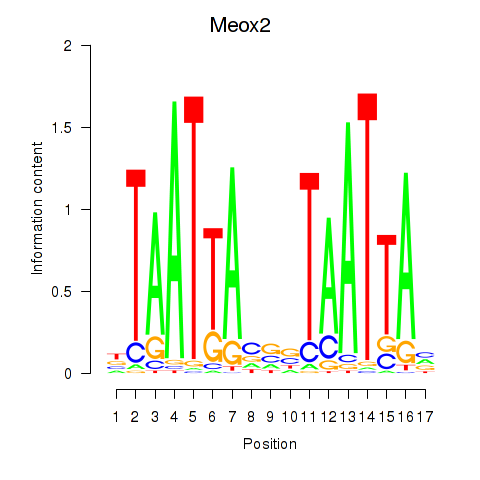
Results for Meox2
Z-value: 1.03
Transcription factors associated with Meox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox2
|
ENSMUSG00000036144.7 | Meox2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meox2 | mm39_v1_chr12_+_37158532_37158545 | 0.51 | 5.4e-06 | Click! |
Activity profile of Meox2 motif
Sorted Z-values of Meox2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Meox2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_68609426 | 9.93 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr6_+_68247469 | 7.22 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr16_-_19079594 | 6.30 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr1_+_174000304 | 5.86 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr17_-_34506744 | 5.61 |
ENSMUST00000174751.2
ENSMUST00000040655.14 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr6_+_41511733 | 4.80 |
ENSMUST00000103287.2
|
Trbj1-4
|
T cell receptor beta joining 1-4 |
| chr18_+_60936910 | 4.79 |
ENSMUST00000097563.9
ENSMUST00000050487.16 ENSMUST00000167610.2 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr6_-_41012435 | 4.75 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr6_+_41510925 | 4.72 |
ENSMUST00000103285.2
|
Trbj1-2
|
T cell receptor beta joining 1-2 |
| chr6_-_69877961 | 4.64 |
ENSMUST00000197290.2
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr17_+_34457868 | 4.57 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr6_+_41512010 | 4.45 |
ENSMUST00000103288.2
|
Trbj1-5
|
T cell receptor beta joining 1-5 |
| chr6_+_41511248 | 4.07 |
ENSMUST00000192366.2
ENSMUST00000103286.2 |
Trbj1-3
|
T cell receptor beta joining 1-3 |
| chr13_-_19491721 | 4.05 |
ENSMUST00000103561.3
|
Trgc2
|
T cell receptor gamma, constant 2 |
| chr10_-_88518878 | 3.97 |
ENSMUST00000004473.15
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr6_+_68098030 | 3.92 |
ENSMUST00000103317.3
|
Igkv1-117
|
immunoglobulin kappa variable 1-117 |
| chr13_+_19398273 | 3.76 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chr14_-_61495934 | 3.71 |
ENSMUST00000077954.13
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr6_+_70584375 | 3.63 |
ENSMUST00000197560.2
|
Igkv3-7
|
immunoglobulin kappa variable 3-7 |
| chr11_+_46701619 | 3.46 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr6_-_68713748 | 3.46 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr13_+_19445054 | 3.38 |
ENSMUST00000164407.2
ENSMUST00000198163.2 |
Trgc3
|
T cell receptor gamma, constant 3 |
| chr14_-_61495832 | 3.31 |
ENSMUST00000121148.8
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr12_+_108572015 | 3.12 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr1_-_194859015 | 3.10 |
ENSMUST00000082321.9
ENSMUST00000195120.6 |
Cr2
|
complement receptor 2 |
| chr6_+_68196928 | 3.10 |
ENSMUST00000103318.6
ENSMUST00000103319.3 |
Igkv2-112
|
immunoglobulin kappa variable 2-112 |
| chr12_-_114621406 | 3.04 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr4_+_114914880 | 2.99 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr7_-_3901119 | 2.88 |
ENSMUST00000070639.8
|
Gm14548
|
predicted gene 14548 |
| chr1_-_194858918 | 2.84 |
ENSMUST00000210219.2
ENSMUST00000193801.2 |
Cr2
|
complement receptor 2 |
| chr6_+_41510788 | 2.80 |
ENSMUST00000103284.2
|
Trbj1-1
|
T cell receptor beta joining 1-1 |
| chr3_-_59009231 | 2.64 |
ENSMUST00000085040.5
|
Gpr171
|
G protein-coupled receptor 171 |
| chr10_-_81335966 | 2.61 |
ENSMUST00000053646.7
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr5_-_105198913 | 2.60 |
ENSMUST00000112718.5
|
Gbp8
|
guanylate-binding protein 8 |
| chr16_+_57369595 | 2.50 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr5_-_108943211 | 2.46 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr14_-_20706556 | 2.44 |
ENSMUST00000090469.8
|
Myoz1
|
myozenin 1 |
| chr17_+_87270504 | 2.24 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr7_-_30523191 | 2.23 |
ENSMUST00000053156.10
|
Ffar2
|
free fatty acid receptor 2 |
| chrX_+_56257374 | 2.22 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr15_+_103148824 | 2.20 |
ENSMUST00000036004.16
ENSMUST00000087351.9 ENSMUST00000231141.2 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr18_-_43925932 | 2.19 |
ENSMUST00000237926.2
ENSMUST00000096570.4 |
Gm94
|
predicted gene 94 |
| chr6_-_78355834 | 2.18 |
ENSMUST00000089667.8
ENSMUST00000167492.4 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr6_-_69282389 | 2.17 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr1_-_69726384 | 2.11 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr17_+_75312520 | 2.10 |
ENSMUST00000234490.2
ENSMUST00000001927.12 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr15_-_99549457 | 2.10 |
ENSMUST00000171908.2
ENSMUST00000171702.8 ENSMUST00000109581.3 ENSMUST00000169810.8 ENSMUST00000023756.12 |
Racgap1
|
Rac GTPase-activating protein 1 |
| chr14_-_20546848 | 2.07 |
ENSMUST00000022353.5
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr6_+_41279199 | 2.06 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr1_-_173301492 | 1.96 |
ENSMUST00000169797.2
|
Ifi206
|
interferon activated gene 206 |
| chr6_+_70495224 | 1.96 |
ENSMUST00000103396.2
|
Igkv3-12
|
immunoglobulin kappa variable 3-12 |
| chr4_-_136626073 | 1.94 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr12_+_117807224 | 1.94 |
ENSMUST00000021592.16
|
Cdca7l
|
cell division cycle associated 7 like |
| chr4_+_66745803 | 1.88 |
ENSMUST00000048096.12
ENSMUST00000107365.3 |
Tlr4
|
toll-like receptor 4 |
| chr12_-_114878652 | 1.83 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr7_+_139827152 | 1.83 |
ENSMUST00000164583.8
ENSMUST00000093984.3 |
Scart2
|
scavenger receptor family member expressed on T cells 2 |
| chr12_-_115798038 | 1.82 |
ENSMUST00000103544.3
|
Ighv1-75
|
immunoglobulin heavy variable 1-75 |
| chr12_-_114955196 | 1.82 |
ENSMUST00000194865.2
|
Ighv1-47
|
immunoglobulin heavy variable 1-47 |
| chr3_-_98247237 | 1.81 |
ENSMUST00000065793.12
|
Phgdh
|
3-phosphoglycerate dehydrogenase |
| chr5_-_65090893 | 1.81 |
ENSMUST00000197315.5
|
Tlr1
|
toll-like receptor 1 |
| chr9_+_72345267 | 1.79 |
ENSMUST00000183809.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr6_-_69477770 | 1.78 |
ENSMUST00000197448.2
|
Igkv4-58
|
immunoglobulin kappa variable 4-58 |
| chr7_+_27259895 | 1.77 |
ENSMUST00000187032.2
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr7_+_88079465 | 1.69 |
ENSMUST00000107256.4
|
Rab38
|
RAB38, member RAS oncogene family |
| chr16_-_56688024 | 1.66 |
ENSMUST00000232373.2
|
Tmem45a
|
transmembrane protein 45a |
| chr8_+_66838927 | 1.65 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr10_+_102376109 | 1.61 |
ENSMUST00000055355.6
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr8_+_3410842 | 1.60 |
ENSMUST00000238813.2
|
Arhgef18
|
rho/rac guanine nucleotide exchange factor (GEF) 18 |
| chr13_+_83652352 | 1.57 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr3_-_154760978 | 1.54 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr7_-_67913949 | 1.51 |
ENSMUST00000032770.4
ENSMUST00000207874.2 |
Pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr15_+_102204691 | 1.50 |
ENSMUST00000064924.6
ENSMUST00000230212.2 ENSMUST00000229050.2 ENSMUST00000231104.2 |
Espl1
|
extra spindle pole bodies 1, separase |
| chr2_+_87696836 | 1.49 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr4_+_134658209 | 1.47 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr2_+_32617671 | 1.46 |
ENSMUST00000113242.5
|
Sh2d3c
|
SH2 domain containing 3C |
| chr1_+_189460461 | 1.45 |
ENSMUST00000097442.9
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr12_-_114451189 | 1.43 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr17_-_33937565 | 1.42 |
ENSMUST00000174040.2
ENSMUST00000173015.8 ENSMUST00000066121.13 ENSMUST00000186022.7 ENSMUST00000173329.8 ENSMUST00000172767.9 |
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr7_-_3828640 | 1.41 |
ENSMUST00000189095.7
ENSMUST00000094911.5 ENSMUST00000153846.8 ENSMUST00000108619.8 ENSMUST00000108620.8 |
Gm15448
|
predicted gene 15448 |
| chr15_+_100659622 | 1.41 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr11_-_69556888 | 1.41 |
ENSMUST00000108654.3
|
Cd68
|
CD68 antigen |
| chr3_-_72875187 | 1.37 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr13_+_28441511 | 1.36 |
ENSMUST00000223428.2
|
Rps18-ps5
|
ribosomal protein S18, pseudogene 5 |
| chr5_-_147831610 | 1.34 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr6_+_47897410 | 1.32 |
ENSMUST00000009411.9
|
Zfp212
|
Zinc finger protein 212 |
| chr1_-_144427302 | 1.32 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr2_+_103254401 | 1.31 |
ENSMUST00000028609.14
|
Elf5
|
E74-like factor 5 |
| chr8_-_125675901 | 1.31 |
ENSMUST00000034469.7
|
Egln1
|
egl-9 family hypoxia-inducible factor 1 |
| chr1_-_106980033 | 1.29 |
ENSMUST00000112717.3
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr16_-_16494269 | 1.29 |
ENSMUST00000206365.3
|
Olfr19
|
olfactory receptor 19 |
| chr2_-_17465410 | 1.29 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr14_+_53007210 | 1.29 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr6_+_67993691 | 1.29 |
ENSMUST00000103314.3
|
Igkv1-122
|
immunoglobulin kappa chain variable 1-122 |
| chr3_+_115681486 | 1.28 |
ENSMUST00000189799.7
ENSMUST00000200258.2 |
Dph5
|
diphthamide biosynthesis 5 |
| chr2_+_103788321 | 1.28 |
ENSMUST00000156813.8
ENSMUST00000170926.8 |
Lmo2
|
LIM domain only 2 |
| chr9_-_36637670 | 1.27 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr9_+_94551929 | 1.25 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr6_-_71417607 | 1.25 |
ENSMUST00000002292.15
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr11_+_46345784 | 1.20 |
ENSMUST00000109229.2
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr13_+_104365880 | 1.16 |
ENSMUST00000022227.8
|
Cenpk
|
centromere protein K |
| chr14_+_73376192 | 1.13 |
ENSMUST00000171070.8
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr14_+_54082691 | 1.11 |
ENSMUST00000103674.6
|
Trav19
|
T cell receptor alpha variable 19 |
| chr19_-_34143437 | 1.09 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr12_-_55033130 | 1.09 |
ENSMUST00000173433.8
ENSMUST00000173803.2 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr13_+_83652280 | 1.09 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr9_-_36637923 | 1.08 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr2_+_89821565 | 1.07 |
ENSMUST00000111509.3
ENSMUST00000213909.3 |
Olfr1261
|
olfactory receptor 1261 |
| chr11_+_46345747 | 1.06 |
ENSMUST00000020668.15
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr10_-_119948890 | 1.05 |
ENSMUST00000020449.12
|
Helb
|
helicase (DNA) B |
| chr7_-_34012956 | 1.04 |
ENSMUST00000108074.8
|
Garre1
|
granule associated Rac and RHOG effector 1 |
| chr3_+_66127330 | 1.04 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr4_+_102278715 | 1.02 |
ENSMUST00000106904.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr12_-_115825934 | 1.02 |
ENSMUST00000198777.2
|
Ighv1-77
|
immunoglobulin heavy variable 1-77 |
| chr14_+_52904815 | 1.02 |
ENSMUST00000198058.2
ENSMUST00000103573.3 |
Trav6-2
|
T cell receptor alpha variable 6-2 |
| chr17_-_40553176 | 1.01 |
ENSMUST00000026499.6
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr14_+_53599724 | 0.99 |
ENSMUST00000196105.2
|
Trav13n-4
|
T cell receptor alpha variable 13N-4 |
| chr7_-_34012934 | 0.98 |
ENSMUST00000206399.2
|
Garre1
|
granule associated Rac and RHOG effector 1 |
| chr6_-_68968278 | 0.92 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr16_-_58620631 | 0.91 |
ENSMUST00000206205.3
|
Olfr173
|
olfactory receptor 173 |
| chr16_+_4457805 | 0.91 |
ENSMUST00000038770.4
|
Vasn
|
vasorin |
| chr10_-_51507527 | 0.89 |
ENSMUST00000219286.2
ENSMUST00000020062.4 ENSMUST00000218684.2 |
Gprc6a
|
G protein-coupled receptor, family C, group 6, member A |
| chr15_-_103148239 | 0.89 |
ENSMUST00000118152.8
|
Cbx5
|
chromobox 5 |
| chr12_-_115410489 | 0.89 |
ENSMUST00000194581.2
|
Ighv1-62-2
|
immunoglobulin heavy variable 1-62-2 |
| chr9_+_38312994 | 0.88 |
ENSMUST00000214648.3
ENSMUST00000056364.3 |
Olfr147
|
olfactory receptor 147 |
| chr18_+_75133519 | 0.88 |
ENSMUST00000079716.6
|
Rpl17
|
ribosomal protein L17 |
| chr2_+_26471062 | 0.88 |
ENSMUST00000238951.2
ENSMUST00000166920.10 |
Egfl7
|
EGF-like domain 7 |
| chr16_+_33071784 | 0.87 |
ENSMUST00000023502.6
|
Snx4
|
sorting nexin 4 |
| chr4_+_103000248 | 0.86 |
ENSMUST00000106855.2
|
Mier1
|
MEIR1 treanscription regulator |
| chr6_-_87815653 | 0.86 |
ENSMUST00000204431.2
ENSMUST00000089497.7 |
Isy1
|
ISY1 splicing factor homolog |
| chr19_+_34078333 | 0.86 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr4_+_11558905 | 0.85 |
ENSMUST00000095145.12
ENSMUST00000108306.9 ENSMUST00000070755.13 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr8_+_112264095 | 0.85 |
ENSMUST00000173726.8
ENSMUST00000174454.8 |
Znrf1
|
zinc and ring finger 1 |
| chr14_+_53328803 | 0.84 |
ENSMUST00000103609.2
|
Trav7n-4
|
T cell receptor alpha variable 7N-4 |
| chr1_-_155910546 | 0.84 |
ENSMUST00000169241.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr1_+_134333720 | 0.82 |
ENSMUST00000173908.8
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr11_-_99494134 | 0.79 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr10_-_43934774 | 0.78 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr7_+_45271229 | 0.75 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr6_+_41928559 | 0.74 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr10_+_58207229 | 0.73 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr16_-_56891711 | 0.72 |
ENSMUST00000067173.8
ENSMUST00000227043.2 |
Tmem45a2
|
transmembrane protein 45A2 |
| chr2_-_150097511 | 0.72 |
ENSMUST00000063463.6
|
Gm21994
|
predicted gene 21994 |
| chr15_+_74593041 | 0.71 |
ENSMUST00000070923.3
|
Them6
|
thioesterase superfamily member 6 |
| chr6_-_116561156 | 0.70 |
ENSMUST00000061723.6
|
Olfr215
|
olfactory receptor 215 |
| chr2_-_85916198 | 0.70 |
ENSMUST00000099911.5
|
Olfr1037
|
olfactory receptor 1037 |
| chr11_-_71092282 | 0.70 |
ENSMUST00000108515.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr1_-_37955569 | 0.68 |
ENSMUST00000078307.7
|
Lyg2
|
lysozyme G-like 2 |
| chr13_-_23417289 | 0.67 |
ENSMUST00000077116.5
|
Vmn1r222
|
vomeronasal 1 receptor 222 |
| chr14_-_55950545 | 0.66 |
ENSMUST00000002389.9
ENSMUST00000226907.2 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr12_-_114815280 | 0.66 |
ENSMUST00000103512.3
|
Ighv1-34
|
immunoglobulin heavy variable 1-34 |
| chr14_+_50360643 | 0.66 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727 |
| chr2_+_91376650 | 0.64 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr8_-_57003828 | 0.63 |
ENSMUST00000134162.8
ENSMUST00000140107.8 ENSMUST00000040330.15 ENSMUST00000135337.8 |
Cep44
|
centrosomal protein 44 |
| chr12_-_115019136 | 0.63 |
ENSMUST00000103519.2
ENSMUST00000192724.2 |
Ighv1-49
|
immunoglobulin heavy variable 1-49 |
| chr16_+_42727926 | 0.62 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_-_52859227 | 0.62 |
ENSMUST00000107670.3
|
Olfr273
|
olfactory receptor 273 |
| chr3_+_32490300 | 0.62 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr6_+_43212512 | 0.60 |
ENSMUST00000078057.4
|
Olfr47
|
olfactory receptor 47 |
| chr3_+_92874366 | 0.60 |
ENSMUST00000098886.5
|
Lce3e
|
late cornified envelope 3E |
| chr3_-_151899470 | 0.59 |
ENSMUST00000050073.13
|
Dnajb4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr13_-_97897139 | 0.59 |
ENSMUST00000074072.5
|
Rps18-ps6
|
ribosomal protein S18, pseudogene 6 |
| chr7_-_89630141 | 0.58 |
ENSMUST00000238981.2
ENSMUST00000208977.2 ENSMUST00000107234.3 |
Eed
|
embryonic ectoderm development |
| chr4_-_144447974 | 0.57 |
ENSMUST00000036876.8
|
AAdacl4fm3
|
AADACL4 family member 3 |
| chr10_+_25274491 | 0.57 |
ENSMUST00000219900.2
|
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr9_-_20247390 | 0.56 |
ENSMUST00000212943.4
ENSMUST00000086473.4 |
Olfr18
|
olfactory receptor 18 |
| chr8_-_46747629 | 0.56 |
ENSMUST00000058636.9
|
Helt
|
helt bHLH transcription factor |
| chr9_+_20209828 | 0.54 |
ENSMUST00000215540.2
ENSMUST00000075717.7 |
Olfr873
|
olfactory receptor 873 |
| chr11_+_118324652 | 0.52 |
ENSMUST00000106286.3
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr7_-_139628991 | 0.51 |
ENSMUST00000172775.4
ENSMUST00000055353.9 |
Msx3
|
msh homeobox 3 |
| chr5_-_109372574 | 0.51 |
ENSMUST00000170341.3
|
Vmn2r14
|
vomeronasal 2, receptor 14 |
| chr13_-_30039613 | 0.51 |
ENSMUST00000091674.12
ENSMUST00000006353.14 |
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr11_-_74243447 | 0.50 |
ENSMUST00000141134.2
ENSMUST00000214769.2 |
Olfr411
|
olfactory receptor 411 |
| chr4_-_112291169 | 0.49 |
ENSMUST00000058605.3
|
Skint9
|
selection and upkeep of intraepithelial T cells 9 |
| chrX_+_41578568 | 0.49 |
ENSMUST00000197237.2
|
Tex13c2
|
TEX13 family member C2 |
| chr1_-_155910567 | 0.49 |
ENSMUST00000141878.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr6_+_132928065 | 0.48 |
ENSMUST00000070991.5
|
Tas2r129
|
taste receptor, type 2, member 129 |
| chr6_+_5390386 | 0.48 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr1_+_178233640 | 0.47 |
ENSMUST00000027775.9
|
Efcab2
|
EF-hand calcium binding domain 2 |
| chr2_+_120459593 | 0.47 |
ENSMUST00000180041.9
|
Stard9
|
START domain containing 9 |
| chr19_-_12219023 | 0.46 |
ENSMUST00000087818.2
|
Olfr262
|
olfactory receptor 262 |
| chr7_-_5808444 | 0.46 |
ENSMUST00000075085.7
|
Vmn1r63
|
vomeronasal 1 receptor 63 |
| chr12_-_115567853 | 0.45 |
ENSMUST00000103538.3
ENSMUST00000198646.2 |
Ighv1-67
|
immunoglobulin heavy variable V1-67 |
| chr16_+_65317389 | 0.45 |
ENSMUST00000176330.8
ENSMUST00000004964.15 ENSMUST00000176038.8 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr16_-_55659194 | 0.44 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr16_-_44379226 | 0.44 |
ENSMUST00000114634.3
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
| chr7_-_66915756 | 0.44 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr9_+_38164715 | 0.44 |
ENSMUST00000093865.3
|
Olfr143
|
olfactory receptor 143 |
| chr11_-_58059293 | 0.42 |
ENSMUST00000172035.8
ENSMUST00000035604.13 ENSMUST00000102711.9 |
Gemin5
|
gem nuclear organelle associated protein 5 |
| chr4_-_75196485 | 0.42 |
ENSMUST00000030103.9
|
Dmac1
|
distal membrane arm assembly complex 1 |
| chr1_+_92500847 | 0.41 |
ENSMUST00000074859.3
|
Olfr1413
|
olfactory receptor 1413 |
| chr15_+_31224555 | 0.40 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr8_+_34007333 | 0.40 |
ENSMUST00000124496.8
|
Tex15
|
testis expressed gene 15 |
| chr2_-_37007795 | 0.40 |
ENSMUST00000213817.2
ENSMUST00000215927.2 |
Olfr362
|
olfactory receptor 362 |
| chr11_+_113575208 | 0.40 |
ENSMUST00000042227.15
ENSMUST00000123466.2 ENSMUST00000106621.4 |
D11Wsu47e
|
DNA segment, Chr 11, Wayne State University 47, expressed |
| chr2_-_88157559 | 0.40 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr5_+_5623799 | 0.39 |
ENSMUST00000101627.5
|
Gm8773
|
predicted gene 8773 |
| chr9_+_38164070 | 0.38 |
ENSMUST00000213129.2
|
Olfr143
|
olfactory receptor 143 |
| chr8_+_34006758 | 0.38 |
ENSMUST00000149399.8
|
Tex15
|
testis expressed gene 15 |
| chr5_-_86893645 | 0.38 |
ENSMUST00000161306.2
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr9_+_64142483 | 0.38 |
ENSMUST00000039011.4
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.0 | 3.0 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.0 | 4.8 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.8 | 2.3 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.7 | 2.2 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.6 | 1.9 | GO:0045362 | nitric oxide production involved in inflammatory response(GO:0002537) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.6 | 1.8 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) |
| 0.6 | 2.3 | GO:2000521 | negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.6 | 2.2 | GO:0002879 | cell surface pattern recognition receptor signaling pathway(GO:0002752) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.4 | 1.7 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.4 | 1.8 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.3 | 1.0 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.3 | 1.3 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 5.6 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.3 | 0.9 | GO:0000389 | generation of catalytic spliceosome for second transesterification step(GO:0000350) mRNA 3'-splice site recognition(GO:0000389) |
| 0.3 | 1.1 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.3 | 1.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 1.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 1.7 | GO:0090383 | platelet dense granule organization(GO:0060155) phagosome acidification(GO:0090383) |
| 0.2 | 2.7 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 3.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 0.6 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 0.6 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.2 | 1.3 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.2 | 1.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 21.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 5.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 1.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.4 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 27.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.9 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.9 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 2.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 1.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.7 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 1.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.5 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 1.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.7 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 2.1 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.9 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 1.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.7 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 0.1 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 2.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.4 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 0.3 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.9 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.7 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:1990166 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.3 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 2.6 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.5 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.5 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.6 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 7.2 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 2.4 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 2.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 2.9 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 1.3 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 1.4 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.0 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 1.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 15.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.0 | 3.0 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.8 | 5.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.8 | 7.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.7 | 2.1 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.6 | 1.8 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.3 | 2.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.3 | 1.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 1.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 1.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 2.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 1.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 1.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 1.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 13.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.6 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 8.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.7 | 2.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.7 | 5.9 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.6 | 1.8 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.6 | 2.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.6 | 1.7 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.5 | 2.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 2.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 1.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 5.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 1.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 0.9 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 4.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 1.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.2 | 2.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 28.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.4 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 2.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 2.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 3.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 4.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.2 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.0 | 0.6 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 7.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.8 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.9 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 1.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 6.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 2.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 1.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 2.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 3.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 3.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 5.7 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 5.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 2.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |