Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Mnt
Z-value: 1.30
Transcription factors associated with Mnt
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mnt
|
ENSMUSG00000000282.13 | Mnt |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mnt | mm39_v1_chr11_+_74721733_74721746 | 0.71 | 4.7e-12 | Click! |
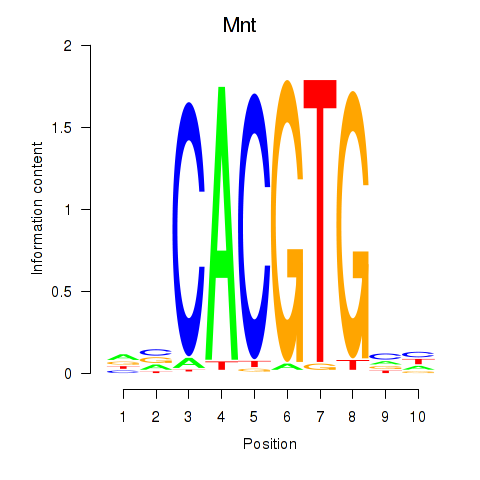
Activity profile of Mnt motif
Sorted Z-values of Mnt motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Mnt
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_151922936 | 25.02 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chr9_-_121324744 | 15.02 |
ENSMUST00000035120.6
|
Cck
|
cholecystokinin |
| chr5_-_148931957 | 14.80 |
ENSMUST00000147473.6
|
Gm42791
|
predicted gene 42791 |
| chr12_-_11486544 | 14.62 |
ENSMUST00000072299.7
|
Vsnl1
|
visinin-like 1 |
| chr4_-_148123223 | 13.34 |
ENSMUST00000030879.12
ENSMUST00000137724.8 |
Clcn6
|
chloride channel, voltage-sensitive 6 |
| chr17_-_56783376 | 13.20 |
ENSMUST00000223859.2
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr17_-_25789652 | 13.18 |
ENSMUST00000025003.10
ENSMUST00000173447.2 |
Sox8
|
SRY (sex determining region Y)-box 8 |
| chr9_-_121324705 | 13.08 |
ENSMUST00000216138.2
|
Cck
|
cholecystokinin |
| chr1_-_22031718 | 12.94 |
ENSMUST00000029667.13
ENSMUST00000173058.8 ENSMUST00000173404.2 |
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr17_-_56783462 | 12.63 |
ENSMUST00000067538.6
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr4_+_134195631 | 12.51 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr9_+_21279161 | 12.26 |
ENSMUST00000067646.12
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr11_+_69231274 | 11.75 |
ENSMUST00000129321.2
|
Rnf227
|
ring finger protein 227 |
| chr9_+_21279179 | 11.14 |
ENSMUST00000213518.2
ENSMUST00000216892.2 |
Ilf3
|
interleukin enhancer binding factor 3 |
| chr12_-_90705212 | 10.75 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr7_+_87233554 | 10.70 |
ENSMUST00000125009.9
|
Grm5
|
glutamate receptor, metabotropic 5 |
| chr9_-_57513510 | 10.69 |
ENSMUST00000215487.2
ENSMUST00000045068.10 |
Cplx3
|
complexin 3 |
| chr18_+_67597929 | 10.57 |
ENSMUST00000025411.9
|
Prelid3a
|
PRELI domain containing 3A |
| chr3_-_95789505 | 10.34 |
ENSMUST00000159863.2
ENSMUST00000159739.8 ENSMUST00000036418.10 ENSMUST00000161866.8 |
Ciart
|
circadian associated repressor of transcription |
| chr5_-_136199525 | 10.30 |
ENSMUST00000041048.6
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr9_-_83688294 | 10.18 |
ENSMUST00000034796.14
ENSMUST00000183614.2 |
Elovl4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 |
| chr9_+_36743980 | 10.11 |
ENSMUST00000034630.15
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr4_+_57568144 | 9.83 |
ENSMUST00000102904.10
|
Pakap
|
paralemmin A kinase anchor protein |
| chrX_+_165127688 | 9.68 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr11_+_69231589 | 9.63 |
ENSMUST00000218008.2
ENSMUST00000151617.3 |
Rnf227
|
ring finger protein 227 |
| chr12_+_16703383 | 9.42 |
ENSMUST00000221596.2
ENSMUST00000111064.3 ENSMUST00000220892.2 |
Ntsr2
|
neurotensin receptor 2 |
| chr17_-_90763300 | 9.27 |
ENSMUST00000159778.8
ENSMUST00000174337.8 ENSMUST00000172466.8 |
Nrxn1
|
neurexin I |
| chr1_-_79836344 | 9.24 |
ENSMUST00000027467.11
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr15_-_102419115 | 9.15 |
ENSMUST00000171565.8
|
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr3_-_57755500 | 9.02 |
ENSMUST00000066882.10
|
Pfn2
|
profilin 2 |
| chr5_-_136199482 | 8.97 |
ENSMUST00000196454.5
ENSMUST00000197052.2 |
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chrX_+_135039745 | 8.96 |
ENSMUST00000116527.2
|
Bex4
|
brain expressed X-linked 4 |
| chr10_-_80837173 | 8.95 |
ENSMUST00000099462.8
ENSMUST00000118233.8 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chrX_-_135116192 | 8.67 |
ENSMUST00000113120.2
ENSMUST00000113118.2 ENSMUST00000058125.9 |
Bex1
|
brain expressed X-linked 1 |
| chr5_-_118382926 | 8.43 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chrX_+_165021919 | 8.34 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr10_+_103203552 | 8.34 |
ENSMUST00000179636.3
ENSMUST00000217905.2 ENSMUST00000074204.12 |
Slc6a15
|
solute carrier family 6 (neurotransmitter transporter), member 15 |
| chrX_-_149800247 | 8.16 |
ENSMUST00000112691.9
ENSMUST00000026297.12 ENSMUST00000154393.8 ENSMUST00000156233.2 |
Gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr18_+_35686424 | 8.07 |
ENSMUST00000235679.2
ENSMUST00000235176.2 ENSMUST00000235801.2 ENSMUST00000237592.2 ENSMUST00000237230.2 ENSMUST00000237589.2 |
Snhg4
Snhg4
|
small nucleolar RNA host gene 4 small nucleolar RNA host gene 4 |
| chr7_+_16609227 | 7.99 |
ENSMUST00000108493.3
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr9_-_35469818 | 7.84 |
ENSMUST00000034612.7
|
Ddx25
|
DEAD box helicase 25 |
| chr1_+_5153300 | 7.84 |
ENSMUST00000044369.13
ENSMUST00000194676.6 ENSMUST00000192029.6 ENSMUST00000192698.3 |
Atp6v1h
|
ATPase, H+ transporting, lysosomal V1 subunit H |
| chr5_+_14075281 | 7.83 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr17_+_87415049 | 7.83 |
ENSMUST00000041369.8
ENSMUST00000234803.2 |
Socs5
|
suppressor of cytokine signaling 5 |
| chr11_+_80367839 | 7.80 |
ENSMUST00000053413.12
ENSMUST00000147694.2 |
Cdk5r1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr14_-_124914516 | 7.78 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chr8_+_120173458 | 7.77 |
ENSMUST00000098363.10
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr3_+_28835425 | 7.56 |
ENSMUST00000060500.9
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr17_-_74017410 | 7.44 |
ENSMUST00000112591.3
ENSMUST00000024858.12 |
Galnt14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr8_+_123844090 | 7.35 |
ENSMUST00000037900.9
|
Cpne7
|
copine VII |
| chrX_-_50770580 | 7.30 |
ENSMUST00000088172.12
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr1_+_181180183 | 7.25 |
ENSMUST00000161880.8
ENSMUST00000027795.14 |
Cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr11_+_103540391 | 7.13 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chr11_+_68986043 | 7.08 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr16_+_94171477 | 6.98 |
ENSMUST00000117648.9
ENSMUST00000147352.8 ENSMUST00000150346.8 ENSMUST00000155692.8 ENSMUST00000153988.9 ENSMUST00000139513.9 ENSMUST00000141856.8 ENSMUST00000152117.8 ENSMUST00000150097.8 ENSMUST00000122895.8 ENSMUST00000151770.8 ENSMUST00000231569.2 ENSMUST00000147046.8 ENSMUST00000149885.8 ENSMUST00000127667.8 ENSMUST00000119131.3 ENSMUST00000145883.2 |
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr5_-_123711098 | 6.92 |
ENSMUST00000031388.13
|
Vps33a
|
VPS33A CORVET/HOPS core subunit |
| chr3_-_107993906 | 6.90 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr16_+_17307464 | 6.86 |
ENSMUST00000115685.10
|
Aifm3
|
apoptosis-inducing factor, mitochondrion-associated 3 |
| chr2_+_92745426 | 6.85 |
ENSMUST00000028648.3
|
Syt13
|
synaptotagmin XIII |
| chr12_+_16703709 | 6.84 |
ENSMUST00000221049.2
|
Ntsr2
|
neurotensin receptor 2 |
| chr19_-_58849407 | 6.83 |
ENSMUST00000066285.6
|
Hspa12a
|
heat shock protein 12A |
| chr5_+_137059127 | 6.80 |
ENSMUST00000041543.9
ENSMUST00000186451.2 |
Vgf
|
VGF nerve growth factor inducible |
| chr5_-_122917341 | 6.76 |
ENSMUST00000198257.5
ENSMUST00000199599.2 ENSMUST00000196742.2 ENSMUST00000200109.5 ENSMUST00000111668.8 |
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr5_+_107585774 | 6.69 |
ENSMUST00000162298.4
ENSMUST00000094541.4 ENSMUST00000211896.2 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr9_-_113855776 | 6.67 |
ENSMUST00000035090.14
|
Fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr1_-_175319842 | 6.65 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chrX_+_165021897 | 6.64 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chr14_-_78970160 | 6.64 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr16_+_17307503 | 6.60 |
ENSMUST00000023448.15
|
Aifm3
|
apoptosis-inducing factor, mitochondrion-associated 3 |
| chr9_+_21279299 | 6.59 |
ENSMUST00000214852.2
ENSMUST00000115414.3 |
Ilf3
|
interleukin enhancer binding factor 3 |
| chr2_+_83642910 | 6.55 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr16_-_43959993 | 6.53 |
ENSMUST00000137557.8
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr7_-_82297676 | 6.46 |
ENSMUST00000207693.2
ENSMUST00000056728.5 ENSMUST00000156720.8 ENSMUST00000126478.2 |
Saxo2
|
stabilizer of axonemal microtubules 2 |
| chr9_+_36744016 | 6.45 |
ENSMUST00000214772.2
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr10_-_79975181 | 6.41 |
ENSMUST00000105369.8
|
Cbarp
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein |
| chrX_+_80114242 | 6.31 |
ENSMUST00000171953.8
ENSMUST00000026760.3 |
Tmem47
|
transmembrane protein 47 |
| chr15_-_79718423 | 6.29 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr3_+_115801869 | 6.29 |
ENSMUST00000106502.2
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr4_+_32657105 | 6.21 |
ENSMUST00000071642.11
ENSMUST00000178134.2 |
Mdn1
|
midasin AAA ATPase 1 |
| chr7_-_62862261 | 6.15 |
ENSMUST00000032738.7
|
Chrna7
|
cholinergic receptor, nicotinic, alpha polypeptide 7 |
| chrX_-_135104589 | 6.10 |
ENSMUST00000066819.11
|
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr8_-_55177352 | 6.08 |
ENSMUST00000129132.3
ENSMUST00000150488.8 ENSMUST00000127511.9 |
Wdr17
|
WD repeat domain 17 |
| chr8_+_59365291 | 6.04 |
ENSMUST00000160055.2
|
BC030500
|
cDNA sequence BC030500 |
| chr11_-_6150411 | 6.02 |
ENSMUST00000066496.10
|
Nudcd3
|
NudC domain containing 3 |
| chr11_+_87018079 | 5.96 |
ENSMUST00000139532.2
|
Trim37
|
tripartite motif-containing 37 |
| chr15_+_82159094 | 5.96 |
ENSMUST00000116423.3
ENSMUST00000230418.2 |
Septin3
|
septin 3 |
| chr4_-_140781015 | 5.96 |
ENSMUST00000102491.10
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr11_+_87017878 | 5.96 |
ENSMUST00000041282.13
|
Trim37
|
tripartite motif-containing 37 |
| chr5_+_63806451 | 5.95 |
ENSMUST00000159584.3
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr8_+_85763780 | 5.92 |
ENSMUST00000211601.2
ENSMUST00000166592.2 |
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr13_-_48779072 | 5.91 |
ENSMUST00000035824.11
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr14_-_119162736 | 5.87 |
ENSMUST00000004055.10
|
Dzip1
|
DAZ interacting protein 1 |
| chr4_+_148123490 | 5.87 |
ENSMUST00000097788.11
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr16_-_20440005 | 5.81 |
ENSMUST00000052939.4
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr19_-_4251589 | 5.79 |
ENSMUST00000237923.2
ENSMUST00000025740.8 ENSMUST00000237723.2 |
Rad9a
|
RAD9 checkpoint clamp component A |
| chr16_+_10884156 | 5.75 |
ENSMUST00000089011.6
|
Snn
|
stannin |
| chr3_+_65016750 | 5.70 |
ENSMUST00000049230.11
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr4_-_140780972 | 5.69 |
ENSMUST00000040222.14
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr4_-_133484080 | 5.67 |
ENSMUST00000008024.7
|
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr15_-_44651411 | 5.66 |
ENSMUST00000090057.6
ENSMUST00000110269.8 ENSMUST00000228639.2 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chr13_+_104246259 | 5.60 |
ENSMUST00000160322.8
ENSMUST00000159574.2 |
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr15_+_87428483 | 5.59 |
ENSMUST00000230414.2
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr7_-_44635740 | 5.58 |
ENSMUST00000209056.3
ENSMUST00000209124.2 ENSMUST00000208312.2 ENSMUST00000207659.2 ENSMUST00000045325.14 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr15_+_82159398 | 5.53 |
ENSMUST00000023095.14
ENSMUST00000230365.2 |
Septin3
|
septin 3 |
| chrX_+_152615221 | 5.49 |
ENSMUST00000148708.2
ENSMUST00000123264.2 ENSMUST00000049999.9 |
Spin2c
|
spindlin family, member 2C |
| chr13_+_104246245 | 5.48 |
ENSMUST00000044385.14
|
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr3_-_57754901 | 5.45 |
ENSMUST00000120289.3
|
Pfn2
|
profilin 2 |
| chr16_-_43960045 | 5.44 |
ENSMUST00000147025.2
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr9_+_58489523 | 5.42 |
ENSMUST00000177292.8
ENSMUST00000085651.12 ENSMUST00000176557.8 ENSMUST00000114121.11 ENSMUST00000177064.8 |
Nptn
|
neuroplastin |
| chrX_-_149879501 | 5.36 |
ENSMUST00000112683.9
ENSMUST00000026295.10 |
Tsr2
|
TSR2 20S rRNA accumulation |
| chr11_+_60244132 | 5.36 |
ENSMUST00000070805.13
ENSMUST00000094140.9 ENSMUST00000108723.9 ENSMUST00000108722.5 |
Drc3
|
dynein regulatory complex subunit 3 |
| chr6_-_77956635 | 5.34 |
ENSMUST00000161846.8
ENSMUST00000160894.8 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr15_-_43034205 | 5.33 |
ENSMUST00000063492.8
ENSMUST00000226810.2 |
Rspo2
|
R-spondin 2 |
| chr12_+_4183394 | 5.30 |
ENSMUST00000152065.8
ENSMUST00000127756.8 |
Adcy3
|
adenylate cyclase 3 |
| chr2_+_25318642 | 5.30 |
ENSMUST00000102919.4
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr2_-_35869636 | 5.27 |
ENSMUST00000028248.11
ENSMUST00000112976.9 |
Ttll11
|
tubulin tyrosine ligase-like family, member 11 |
| chr3_+_107186151 | 5.27 |
ENSMUST00000145735.6
|
Lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr16_-_43959361 | 5.24 |
ENSMUST00000124102.8
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr1_+_166081755 | 5.21 |
ENSMUST00000194964.6
ENSMUST00000192638.6 ENSMUST00000192426.6 ENSMUST00000195557.6 ENSMUST00000192732.6 ENSMUST00000193860.2 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr3_-_89153135 | 5.20 |
ENSMUST00000041022.15
|
Trim46
|
tripartite motif-containing 46 |
| chr4_-_110209106 | 5.18 |
ENSMUST00000106603.9
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr6_-_77956499 | 5.14 |
ENSMUST00000159626.8
ENSMUST00000075340.12 ENSMUST00000162273.2 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr17_-_50600620 | 5.13 |
ENSMUST00000010736.9
|
Dazl
|
deleted in azoospermia-like |
| chr12_+_102094977 | 5.10 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr8_+_85763534 | 5.10 |
ENSMUST00000093360.12
|
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr6_+_141470105 | 5.09 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr13_-_54209669 | 5.01 |
ENSMUST00000021932.6
ENSMUST00000221470.2 |
Drd1
|
dopamine receptor D1 |
| chr5_-_45796857 | 5.00 |
ENSMUST00000016023.9
|
Fam184b
|
family with sequence similarity 184, member B |
| chr2_+_164810139 | 4.92 |
ENSMUST00000202479.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr15_+_38940307 | 4.79 |
ENSMUST00000067072.5
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chrX_+_5959507 | 4.74 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr9_-_103639018 | 4.71 |
ENSMUST00000215136.2
ENSMUST00000189588.7 |
Tmem108
|
transmembrane protein 108 |
| chr9_-_22300409 | 4.69 |
ENSMUST00000040912.9
|
Anln
|
anillin, actin binding protein |
| chr9_-_123546642 | 4.64 |
ENSMUST00000165754.8
ENSMUST00000026274.14 |
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr11_+_3282424 | 4.63 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr9_-_103638985 | 4.62 |
ENSMUST00000189066.2
|
Tmem108
|
transmembrane protein 108 |
| chr15_+_38940351 | 4.61 |
ENSMUST00000226433.2
ENSMUST00000132192.3 |
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr16_-_43959559 | 4.56 |
ENSMUST00000063661.13
ENSMUST00000114666.9 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr7_-_44635813 | 4.55 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr8_+_106075475 | 4.54 |
ENSMUST00000073149.7
|
Slc9a5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5 |
| chr10_-_81436671 | 4.51 |
ENSMUST00000151858.8
ENSMUST00000142948.8 ENSMUST00000072020.9 |
Tle6
|
transducin-like enhancer of split 6 |
| chr8_+_14961702 | 4.50 |
ENSMUST00000161162.8
ENSMUST00000110800.9 |
Arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr15_-_79718462 | 4.49 |
ENSMUST00000148358.2
|
Cbx6
|
chromobox 6 |
| chr11_+_102080489 | 4.47 |
ENSMUST00000078975.8
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr11_+_102652228 | 4.47 |
ENSMUST00000103081.10
ENSMUST00000068150.7 |
Adam11
|
a disintegrin and metallopeptidase domain 11 |
| chr19_-_58849380 | 4.46 |
ENSMUST00000235263.2
|
Hspa12a
|
heat shock protein 12A |
| chr7_+_3352019 | 4.46 |
ENSMUST00000100301.11
|
Prkcg
|
protein kinase C, gamma |
| chr14_-_47059546 | 4.46 |
ENSMUST00000226937.2
|
Gmfb
|
glia maturation factor, beta |
| chr4_+_148123554 | 4.45 |
ENSMUST00000141283.8
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr6_+_7555053 | 4.42 |
ENSMUST00000090679.9
ENSMUST00000184986.2 |
Tac1
|
tachykinin 1 |
| chr4_+_130202388 | 4.40 |
ENSMUST00000070532.8
|
Fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr8_-_55177510 | 4.37 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17 |
| chr13_-_48779047 | 4.36 |
ENSMUST00000222028.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr3_-_56091096 | 4.34 |
ENSMUST00000029374.8
|
Nbea
|
neurobeachin |
| chr14_-_78866714 | 4.31 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr3_+_115801106 | 4.23 |
ENSMUST00000029575.12
ENSMUST00000106501.8 |
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr16_-_4698148 | 4.23 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr8_-_67140699 | 4.21 |
ENSMUST00000212563.2
ENSMUST00000070810.8 ENSMUST00000211920.2 |
Npy5r
|
neuropeptide Y receptor Y5 |
| chr8_-_122634418 | 4.18 |
ENSMUST00000045557.10
|
Slc7a5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| chr11_-_100650768 | 4.16 |
ENSMUST00000107363.3
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr9_+_45314436 | 4.11 |
ENSMUST00000041005.6
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr19_+_45003670 | 4.10 |
ENSMUST00000236377.2
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr2_-_33020579 | 4.10 |
ENSMUST00000124000.2
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr6_+_141470261 | 4.08 |
ENSMUST00000203140.2
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr12_+_117497478 | 4.07 |
ENSMUST00000220781.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr5_+_22951015 | 4.02 |
ENSMUST00000197992.2
|
Lhfpl3
|
lipoma HMGIC fusion partner-like 3 |
| chr9_-_42035560 | 4.00 |
ENSMUST00000060989.9
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr15_-_53765869 | 3.99 |
ENSMUST00000078673.14
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr2_-_74409225 | 3.96 |
ENSMUST00000134168.2
ENSMUST00000111993.9 ENSMUST00000064503.13 |
Lnpk
|
lunapark, ER junction formation factor |
| chrX_-_135104386 | 3.90 |
ENSMUST00000151592.8
ENSMUST00000131510.2 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr17_+_6320731 | 3.89 |
ENSMUST00000088940.6
|
Tmem181a
|
transmembrane protein 181A |
| chr7_-_45116216 | 3.87 |
ENSMUST00000210392.2
ENSMUST00000211365.2 |
Bax
|
BCL2-associated X protein |
| chr4_+_130297132 | 3.86 |
ENSMUST00000105993.4
|
Nkain1
|
Na+/K+ transporting ATPase interacting 1 |
| chr14_-_78774201 | 3.86 |
ENSMUST00000123853.9
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chr15_+_99590098 | 3.85 |
ENSMUST00000228185.2
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr11_+_102080446 | 3.84 |
ENSMUST00000070334.10
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr7_-_126548671 | 3.84 |
ENSMUST00000106339.2
ENSMUST00000052937.12 |
Asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr11_+_54195006 | 3.83 |
ENSMUST00000108904.10
ENSMUST00000108905.10 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr5_+_107479023 | 3.82 |
ENSMUST00000031215.15
ENSMUST00000112677.10 |
Brdt
|
bromodomain, testis-specific |
| chr5_-_113968483 | 3.79 |
ENSMUST00000100874.6
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr3_+_107186309 | 3.77 |
ENSMUST00000199317.2
|
Lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr2_+_158636727 | 3.76 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr12_+_4183032 | 3.74 |
ENSMUST00000124505.8
|
Adcy3
|
adenylate cyclase 3 |
| chr1_+_50966670 | 3.71 |
ENSMUST00000081851.4
|
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr1_-_131161312 | 3.71 |
ENSMUST00000212202.2
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr2_-_102231208 | 3.71 |
ENSMUST00000102573.8
|
Trim44
|
tripartite motif-containing 44 |
| chr2_-_139908615 | 3.69 |
ENSMUST00000046656.9
ENSMUST00000099304.4 ENSMUST00000110079.9 |
Tasp1
|
taspase, threonine aspartase 1 |
| chr12_+_102095260 | 3.67 |
ENSMUST00000079020.12
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chrX_+_73372664 | 3.67 |
ENSMUST00000004326.4
|
Plxna3
|
plexin A3 |
| chr2_+_163916042 | 3.67 |
ENSMUST00000018353.14
|
Stk4
|
serine/threonine kinase 4 |
| chr13_+_85337521 | 3.66 |
ENSMUST00000022030.11
|
Ccnh
|
cyclin H |
| chr9_-_123546605 | 3.66 |
ENSMUST00000163207.2
|
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr7_-_16121716 | 3.65 |
ENSMUST00000211741.2
ENSMUST00000210999.2 |
Sae1
|
SUMO1 activating enzyme subunit 1 |
| chr10_+_12966532 | 3.65 |
ENSMUST00000121646.8
ENSMUST00000121325.8 ENSMUST00000121766.8 |
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr11_-_118460736 | 3.64 |
ENSMUST00000136551.3
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr1_-_192718064 | 3.64 |
ENSMUST00000215093.2
ENSMUST00000195354.6 |
Syt14
|
synaptotagmin XIV |
| chr5_+_137059522 | 3.63 |
ENSMUST00000187382.2
|
Vgf
|
VGF nerve growth factor inducible |
| chr15_+_73596602 | 3.62 |
ENSMUST00000230177.2
ENSMUST00000163582.9 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr2_-_102230602 | 3.59 |
ENSMUST00000152929.2
|
Trim44
|
tripartite motif-containing 44 |
| chr9_+_72345801 | 3.57 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 25.8 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 4.7 | 4.7 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 4.4 | 13.2 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 3.6 | 10.7 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 3.1 | 24.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 3.1 | 9.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 2.6 | 7.8 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 2.6 | 10.4 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 2.5 | 20.0 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 2.4 | 7.2 | GO:1990117 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 2.4 | 9.4 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 2.3 | 25.0 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 2.3 | 9.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 2.3 | 6.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 2.2 | 15.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 2.1 | 10.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 2.0 | 7.8 | GO:0021586 | pons maturation(GO:0021586) |
| 1.9 | 9.5 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 1.9 | 28.1 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 1.8 | 10.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.7 | 11.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 1.6 | 4.9 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 1.6 | 9.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.5 | 9.3 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 1.5 | 4.6 | GO:1905006 | positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 1.5 | 5.9 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 1.4 | 6.9 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 1.4 | 5.4 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 1.3 | 4.0 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 1.3 | 1.3 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 1.3 | 14.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.3 | 5.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 1.3 | 7.6 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 1.3 | 5.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.2 | 3.7 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 1.2 | 7.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 1.1 | 4.4 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 1.1 | 4.4 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.1 | 4.2 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 1.0 | 8.3 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 1.0 | 4.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 1.0 | 3.0 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 1.0 | 6.8 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 1.0 | 7.8 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.0 | 14.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.0 | 2.9 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.9 | 2.8 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.9 | 9.3 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.9 | 2.8 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.9 | 3.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.9 | 9.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.9 | 9.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.9 | 3.6 | GO:0061055 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) myotome development(GO:0061055) |
| 0.9 | 21.8 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.8 | 8.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.8 | 2.5 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.8 | 7.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.8 | 4.7 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.8 | 3.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.8 | 3.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.8 | 7.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.8 | 2.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.8 | 3.8 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.8 | 5.3 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.8 | 5.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.7 | 4.5 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.7 | 2.9 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.7 | 2.9 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.7 | 2.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.7 | 3.6 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.7 | 6.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.7 | 2.1 | GO:1990164 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.7 | 4.7 | GO:0071543 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.7 | 5.3 | GO:0060437 | lung growth(GO:0060437) |
| 0.7 | 4.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.7 | 2.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.6 | 7.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.6 | 3.8 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.6 | 18.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.6 | 10.5 | GO:0060134 | regulation of synapse structural plasticity(GO:0051823) prepulse inhibition(GO:0060134) |
| 0.6 | 1.8 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.6 | 17.7 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.6 | 3.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.6 | 3.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.6 | 2.3 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.6 | 3.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.6 | 3.9 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.5 | 3.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.5 | 2.7 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.5 | 3.7 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.5 | 11.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.5 | 2.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.5 | 11.0 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.5 | 12.9 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.5 | 9.4 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.5 | 3.4 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.5 | 1.4 | GO:0021666 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.5 | 2.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 10.7 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.4 | 27.6 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.4 | 8.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.4 | 5.3 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 2.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.4 | 1.7 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.4 | 3.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.4 | 1.3 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.4 | 5.8 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.4 | 2.1 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.4 | 4.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 1.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 4.9 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.4 | 0.7 | GO:0003383 | apical constriction(GO:0003383) |
| 0.4 | 2.6 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.4 | 5.4 | GO:0007135 | meiosis II(GO:0007135) |
| 0.4 | 2.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.4 | 6.4 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.4 | 12.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.4 | 8.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.3 | 2.7 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 2.7 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.3 | 5.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.3 | 8.7 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.3 | 0.6 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.3 | 0.9 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.3 | 1.8 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 4.5 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.3 | 2.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.3 | 1.8 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.3 | 1.8 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.3 | 12.8 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.3 | 5.2 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.3 | 3.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 1.7 | GO:0061197 | fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.3 | 1.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 3.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.3 | 1.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.3 | 0.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 0.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 1.6 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.3 | 0.8 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.3 | 2.3 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.3 | 1.3 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.3 | 2.0 | GO:0015824 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.2 | 4.0 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.2 | 6.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.2 | 3.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 1.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.2 | 0.7 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 | 1.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 | 0.9 | GO:1902219 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.2 | 1.4 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.2 | 3.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.2 | 10.4 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.2 | 5.1 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.2 | 2.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 6.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.2 | 0.9 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.2 | 3.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 2.0 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.2 | 3.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 6.1 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.2 | 2.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.2 | 5.7 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 | 7.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 2.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 9.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.2 | 0.6 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 0.8 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 6.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 1.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 2.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 17.5 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.2 | 7.7 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 0.9 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 5.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 0.7 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 7.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 2.6 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.2 | 2.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.2 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 0.7 | GO:0006272 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.2 | 6.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 0.8 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.2 | 6.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.2 | 13.5 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.2 | 1.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 1.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 6.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 15.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 3.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 1.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 6.9 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 2.1 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 2.9 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 3.5 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 1.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 1.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 1.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 7.8 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 1.8 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 23.9 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.1 | 2.9 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 0.1 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.1 | 1.0 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 3.6 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 3.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 4.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 3.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.5 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 0.7 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 4.7 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 4.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 5.8 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 | 3.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 2.0 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 2.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 2.5 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 1.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 3.2 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 2.3 | GO:0050650 | chondroitin sulfate metabolic process(GO:0030204) chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 4.7 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.4 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 2.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 2.9 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 0.6 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.7 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 1.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 1.0 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 | 1.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 6.9 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 1.7 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.1 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.1 | 4.1 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 2.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 4.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.6 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 2.6 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 3.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 2.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 2.1 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 1.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 1.6 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 1.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 8.8 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.1 | 6.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 2.2 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.8 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.1 | 6.1 | GO:0010771 | negative regulation of cell morphogenesis involved in differentiation(GO:0010771) |
| 0.1 | 0.9 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 4.5 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 1.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 3.2 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.2 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.8 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 1.0 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.9 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 1.7 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 1.9 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 1.6 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.8 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.3 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 1.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.7 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.4 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 4.6 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.0 | 0.6 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.3 | GO:0018298 | rhodopsin mediated signaling pathway(GO:0016056) protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.0 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.0 | 1.1 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:1903553 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 1.9 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 25.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 2.6 | 7.9 | GO:0070985 | TFIIK complex(GO:0070985) |
| 2.5 | 29.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 2.4 | 7.2 | GO:0097144 | BAX complex(GO:0097144) |
| 2.0 | 7.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.7 | 28.1 | GO:0043203 | axon hillock(GO:0043203) |
| 1.5 | 5.9 | GO:1990794 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 1.4 | 6.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.3 | 5.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.3 | 10.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 1.2 | 5.8 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 1.1 | 9.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 1.0 | 28.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.0 | 2.9 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.9 | 3.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.9 | 2.6 | GO:1990879 | CST complex(GO:1990879) |
| 0.8 | 5.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.8 | 4.9 | GO:0097433 | dense body(GO:0097433) |
| 0.8 | 3.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.8 | 10.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.7 | 3.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.7 | 4.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.7 | 9.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.6 | 5.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 1.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.6 | 4.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 2.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.5 | 3.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 13.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.5 | 3.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.5 | 3.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.5 | 4.6 | GO:0071546 | pi-body(GO:0071546) |
| 0.5 | 0.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.5 | 9.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.5 | 10.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.5 | 3.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.5 | 2.8 | GO:0031523 | Myb complex(GO:0031523) |
| 0.5 | 1.8 | GO:1990037 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.5 | 5.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.4 | 11.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.4 | 2.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.4 | 10.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 9.7 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.4 | 2.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 6.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 5.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 1.8 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.3 | 2.8 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 10.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 3.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 8.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 5.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 36.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 5.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.3 | 3.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 1.5 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.3 | 2.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 2.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 0.8 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.3 | 0.8 | GO:0097361 | CIA complex(GO:0097361) |
| 0.3 | 5.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 3.0 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.3 | 3.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 0.8 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 2.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 3.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 2.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 1.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 7.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 5.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 1.8 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 4.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 1.9 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 0.9 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 4.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 3.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 4.6 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.2 | 2.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 4.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 45.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 2.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 10.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 9.0 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.1 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 3.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 1.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 2.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.5 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 11.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 3.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 7.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 13.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 7.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 2.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 2.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 7.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 9.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 6.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 29.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 0.7 | GO:0034719 | SMN complex(GO:0032797) SMN-Sm protein complex(GO:0034719) |
| 0.1 | 5.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 1.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 3.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.3 | GO:0000802 | transverse filament(GO:0000802) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 3.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 9.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 3.1 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 3.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 5.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 4.4 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 11.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 4.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.9 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 7.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.5 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 1.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.3 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 4.6 | 18.5 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 3.5 | 10.5 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 3.4 | 10.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 2.7 | 10.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 2.6 | 25.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 2.5 | 10.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 2.3 | 6.8 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 2.1 | 8.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.8 | 7.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 1.8 | 10.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.5 | 4.5 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.5 | 8.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 1.3 | 4.0 | GO:0004349 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 1.3 | 9.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.3 | 19.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 1.3 | 5.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 1.2 | 3.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.2 | 5.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.2 | 5.8 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 1.2 | 4.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.1 | 9.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.1 | 39.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 1.0 | 34.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 1.0 | 10.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.0 | 3.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.8 | 3.4 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.8 | 10.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.8 | 4.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.8 | 2.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.8 | 2.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.8 | 3.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.7 | 7.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.7 | 15.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.7 | 3.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.7 | 6.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.7 | 6.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.7 | 2.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.7 | 4.7 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.7 | 3.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.7 | 9.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.6 | 11.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.6 | 3.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.6 | 3.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.6 | 1.8 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.6 | 12.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.6 | 4.2 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.6 | 8.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.6 | 8.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.6 | 1.7 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.5 | 1.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.5 | 4.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.5 | 3.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.5 | 4.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 10.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 9.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.5 | 16.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.5 | 4.5 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.4 | 5.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 4.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 3.0 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.4 | 3.0 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.4 | 1.7 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 2.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 1.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.4 | 2.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.4 | 14.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.4 | 1.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.4 | 2.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.4 | 6.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 9.0 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.3 | 7.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 1.0 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.3 | 3.8 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 0.9 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 9.5 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.3 | 1.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 3.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 14.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 3.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 9.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 13.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 2.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 6.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 9.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.3 | 1.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.3 | 2.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 14.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 3.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.0 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 0.7 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.2 | 3.9 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 2.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 4.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 0.9 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.2 | 1.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 1.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 1.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 5.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 17.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 2.4 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.2 | 7.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 13.3 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.2 | 4.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 2.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 1.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 2.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 4.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 14.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 4.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 7.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 2.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 2.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 0.5 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 2.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 0.5 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.2 | 2.0 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.6 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 3.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 5.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 4.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 8.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 6.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.8 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 14.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.7 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 2.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.4 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 3.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 9.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 5.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 5.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 9.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 5.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 1.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 1.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 11.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.7 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 11.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 2.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 3.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 1.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 4.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 7.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 7.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 1.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 27.1 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 0.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 5.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 2.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 9.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.7 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 4.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 2.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 4.8 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 1.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 3.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 3.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 2.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.9 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 1.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 11.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 2.0 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 5.5 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.0 | 6.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) nerve growth factor binding(GO:0048406) |
| 0.0 | 2.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.0 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 12.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 5.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 8.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 20.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 5.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 9.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 2.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 11.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 11.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 26.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 5.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 7.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 4.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 6.0 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 8.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 9.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 3.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 2.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 2.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 3.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 4.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 6.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.7 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 12.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 11.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 8.5 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 34.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.7 | 10.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.6 | 10.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.5 | 10.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.5 | 9.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.5 | 11.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 1.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.4 | 14.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 6.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.4 | 10.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 25.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 3.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.4 | 10.5 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.3 | 17.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 11.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 4.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 7.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 21.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.3 | 52.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.3 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 3.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 7.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 2.5 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 13.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 5.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 4.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 9.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 5.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.2 | 11.2 | REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.2 | 4.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 2.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 4.0 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 4.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 5.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 5.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 4.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 12.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.8 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 4.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 6.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 8.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 4.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.0 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.8 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 2.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 0.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 3.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 1.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 3.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 6.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |