Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
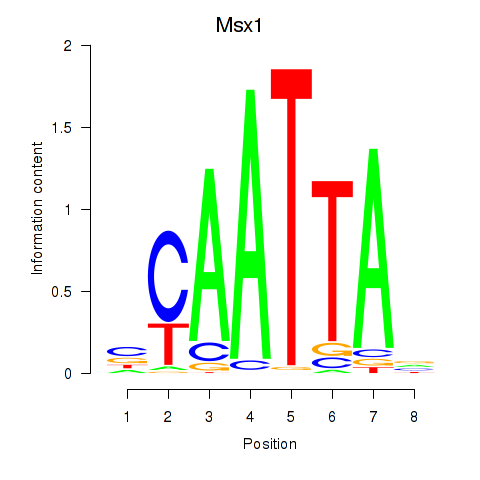
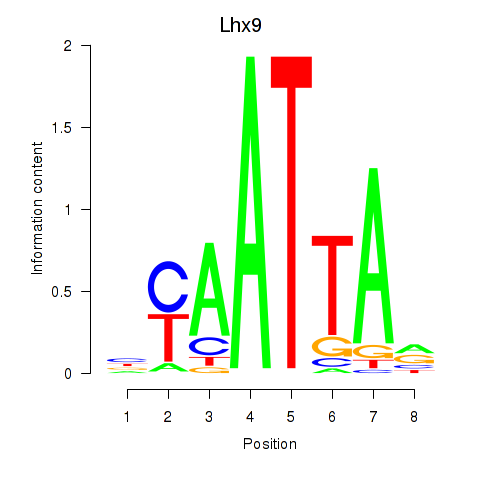
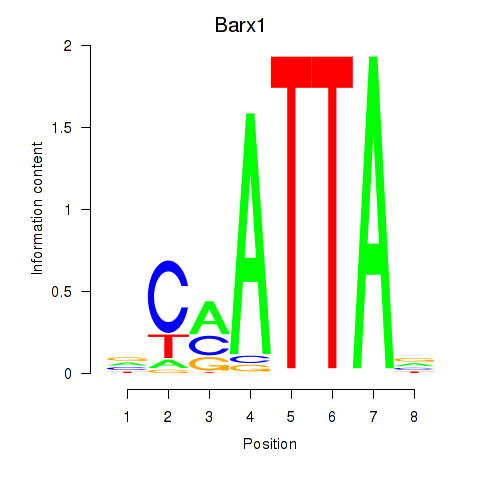
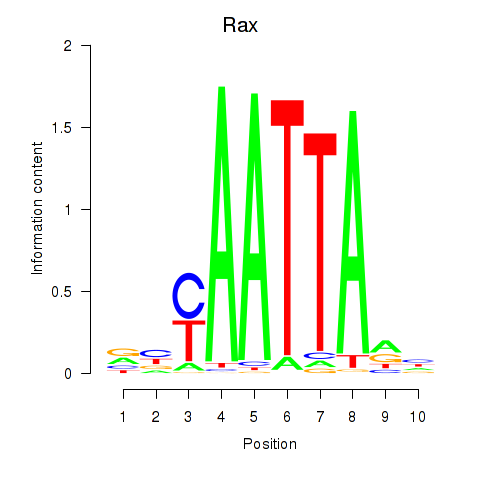
Results for Msx1_Lhx9_Barx1_Rax_Dlx6
Z-value: 0.81
Transcription factors associated with Msx1_Lhx9_Barx1_Rax_Dlx6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx1
|
ENSMUSG00000048450.11 | Msx1 |
|
Lhx9
|
ENSMUSG00000019230.15 | Lhx9 |
|
Barx1
|
ENSMUSG00000021381.5 | Barx1 |
|
Rax
|
ENSMUSG00000024518.5 | Rax |
|
Dlx6
|
ENSMUSG00000029754.14 | Dlx6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rax | mm39_v1_chr18_-_66072168_66072177 | 0.29 | 1.2e-02 | Click! |
| Barx1 | mm39_v1_chr13_+_48816466_48816521 | 0.26 | 2.7e-02 | Click! |
| Lhx9 | mm39_v1_chr1_-_138775317_138775457 | 0.21 | 7.8e-02 | Click! |
| Dlx6 | mm39_v1_chr6_+_6863269_6863334 | 0.10 | 4.1e-01 | Click! |
| Msx1 | mm39_v1_chr5_-_37981919_37981933 | -0.08 | 5.1e-01 | Click! |
Activity profile of Msx1_Lhx9_Barx1_Rax_Dlx6 motif
Sorted Z-values of Msx1_Lhx9_Barx1_Rax_Dlx6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx1_Lhx9_Barx1_Rax_Dlx6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_79417732 | 10.01 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr3_+_93301003 | 7.95 |
ENSMUST00000045912.3
|
Rptn
|
repetin |
| chr15_+_39522905 | 5.92 |
ENSMUST00000226410.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr14_-_110992533 | 5.31 |
ENSMUST00000078386.4
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr2_+_129854256 | 4.97 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr10_-_25076008 | 4.85 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr19_-_24178000 | 4.60 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr5_+_14075281 | 4.54 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr8_+_55407872 | 4.19 |
ENSMUST00000033915.9
|
Gpm6a
|
glycoprotein m6a |
| chr16_-_45544960 | 4.16 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr2_+_27055245 | 4.16 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr2_-_103114105 | 4.03 |
ENSMUST00000111174.8
|
Ehf
|
ets homologous factor |
| chr18_+_23548534 | 3.88 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr9_+_32027335 | 3.75 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr18_+_23548455 | 3.67 |
ENSMUST00000115832.4
|
Dtna
|
dystrobrevin alpha |
| chr16_-_42160957 | 3.60 |
ENSMUST00000102817.5
|
Gap43
|
growth associated protein 43 |
| chr13_+_118851214 | 3.08 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr17_+_12338161 | 3.06 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr13_-_78344492 | 3.05 |
ENSMUST00000125176.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr6_+_15196950 | 2.99 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr3_+_159545309 | 2.95 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr6_-_136758716 | 2.82 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr7_+_126549692 | 2.66 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr2_+_20742115 | 2.66 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr13_-_103042554 | 2.66 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr5_-_134776101 | 2.61 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr10_+_101994841 | 2.60 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr6_+_29859372 | 2.54 |
ENSMUST00000115238.10
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr2_+_3115250 | 2.48 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr17_-_90395771 | 2.47 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr17_-_49871291 | 2.44 |
ENSMUST00000224595.2
ENSMUST00000057610.8 |
Daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr5_-_77262968 | 2.40 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr10_+_53213763 | 2.31 |
ENSMUST00000219491.2
ENSMUST00000163319.9 ENSMUST00000220197.2 ENSMUST00000046221.8 ENSMUST00000218468.2 ENSMUST00000219921.2 |
Pln
|
phospholamban |
| chr6_+_29859660 | 2.20 |
ENSMUST00000128927.9
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr7_-_115797722 | 2.16 |
ENSMUST00000181981.8
|
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr6_+_29859685 | 2.08 |
ENSMUST00000134438.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr5_+_35156389 | 2.08 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr18_-_43925932 | 2.06 |
ENSMUST00000237926.2
ENSMUST00000096570.4 |
Gm94
|
predicted gene 94 |
| chr3_-_49711706 | 2.03 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr12_+_38830283 | 2.01 |
ENSMUST00000162563.8
ENSMUST00000161164.8 ENSMUST00000160996.8 |
Etv1
|
ets variant 1 |
| chr13_-_103042294 | 1.98 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_+_38830812 | 1.85 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr7_+_126550009 | 1.78 |
ENSMUST00000106332.3
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr3_-_49711765 | 1.77 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr10_+_101994719 | 1.76 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr16_-_63684477 | 1.74 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr18_-_38102799 | 1.74 |
ENSMUST00000166148.8
ENSMUST00000163131.8 ENSMUST00000043437.14 |
Fchsd1
|
FCH and double SH3 domains 1 |
| chr11_-_87249837 | 1.69 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr7_+_126549859 | 1.65 |
ENSMUST00000106333.8
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr10_+_73657689 | 1.58 |
ENSMUST00000064562.14
ENSMUST00000193174.6 ENSMUST00000105426.10 ENSMUST00000129404.9 ENSMUST00000131321.9 ENSMUST00000126920.9 ENSMUST00000147189.9 ENSMUST00000105424.10 ENSMUST00000092420.13 ENSMUST00000105429.10 ENSMUST00000193361.6 ENSMUST00000131724.9 ENSMUST00000152655.9 ENSMUST00000151116.9 ENSMUST00000155701.9 ENSMUST00000152819.9 ENSMUST00000125517.9 ENSMUST00000124046.8 ENSMUST00000146682.8 ENSMUST00000177107.8 ENSMUST00000149977.9 ENSMUST00000191854.6 |
Pcdh15
|
protocadherin 15 |
| chr15_+_21111428 | 1.58 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr10_+_39488930 | 1.56 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr18_+_57601541 | 1.55 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr6_+_7555053 | 1.54 |
ENSMUST00000090679.9
ENSMUST00000184986.2 |
Tac1
|
tachykinin 1 |
| chr1_-_158183894 | 1.53 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr8_+_10299288 | 1.51 |
ENSMUST00000214643.2
|
Myo16
|
myosin XVI |
| chr2_+_59442378 | 1.49 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr5_-_62923463 | 1.48 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr7_-_66077215 | 1.48 |
ENSMUST00000015278.15
|
Aldh1a3
|
aldehyde dehydrogenase family 1, subfamily A3 |
| chr16_-_74208180 | 1.44 |
ENSMUST00000117200.8
|
Robo2
|
roundabout guidance receptor 2 |
| chr9_-_40257586 | 1.41 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr3_-_72875187 | 1.41 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr9_+_43222104 | 1.41 |
ENSMUST00000034511.7
|
Trim29
|
tripartite motif-containing 29 |
| chr10_-_85847697 | 1.38 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr12_+_52746158 | 1.37 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr11_-_97944239 | 1.36 |
ENSMUST00000017544.9
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr16_-_74208395 | 1.36 |
ENSMUST00000227347.2
|
Robo2
|
roundabout guidance receptor 2 |
| chr5_+_35156454 | 1.35 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr17_+_17622934 | 1.34 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr1_-_93088562 | 1.34 |
ENSMUST00000143419.2
|
Mab21l4
|
mab-21-like 4 |
| chr5_-_53864874 | 1.34 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr17_-_36008863 | 1.33 |
ENSMUST00000146472.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr14_-_36820304 | 1.33 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr10_-_8632519 | 1.33 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr12_+_38833454 | 1.29 |
ENSMUST00000161980.8
ENSMUST00000160701.8 |
Etv1
|
ets variant 1 |
| chr6_+_125529911 | 1.29 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr4_-_14621805 | 1.27 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr4_-_43710231 | 1.26 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr11_+_115225557 | 1.24 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr17_+_49922129 | 1.22 |
ENSMUST00000162854.2
|
Kif6
|
kinesin family member 6 |
| chr12_+_38833501 | 1.22 |
ENSMUST00000159334.8
|
Etv1
|
ets variant 1 |
| chr15_-_13173736 | 1.20 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chr12_+_31488208 | 1.20 |
ENSMUST00000001254.6
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr12_+_38831093 | 1.17 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr8_-_3675525 | 1.15 |
ENSMUST00000144977.2
ENSMUST00000136105.8 ENSMUST00000128566.8 |
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr7_+_30463175 | 1.12 |
ENSMUST00000165887.8
ENSMUST00000085691.11 ENSMUST00000054427.13 ENSMUST00000085688.11 |
Dmkn
|
dermokine |
| chr19_-_55229668 | 1.12 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr19_+_24651362 | 1.12 |
ENSMUST00000057243.6
|
Tmem252
|
transmembrane protein 252 |
| chr16_-_56688024 | 1.06 |
ENSMUST00000232373.2
|
Tmem45a
|
transmembrane protein 45a |
| chrX_+_158086253 | 1.04 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr10_-_44024843 | 1.04 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr19_-_14575395 | 1.04 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr3_+_68479578 | 1.04 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr16_-_63684425 | 1.00 |
ENSMUST00000232049.2
|
Epha3
|
Eph receptor A3 |
| chr18_+_23548192 | 0.99 |
ENSMUST00000222515.2
|
Dtna
|
dystrobrevin alpha |
| chr5_-_90788323 | 0.94 |
ENSMUST00000202784.4
ENSMUST00000031317.10 ENSMUST00000201370.2 |
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr8_-_62576140 | 0.92 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr1_+_104696235 | 0.91 |
ENSMUST00000062528.9
|
Cdh20
|
cadherin 20 |
| chr3_-_141687987 | 0.90 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr6_-_147165623 | 0.87 |
ENSMUST00000052296.9
ENSMUST00000204197.2 |
Pthlh
|
parathyroid hormone-like peptide |
| chr16_+_13176238 | 0.86 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chr1_-_52230062 | 0.85 |
ENSMUST00000156887.8
ENSMUST00000129107.2 |
Gls
|
glutaminase |
| chr10_+_88721854 | 0.84 |
ENSMUST00000020255.8
|
Slc5a8
|
solute carrier family 5 (iodide transporter), member 8 |
| chr8_+_46111703 | 0.80 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_+_22512195 | 0.79 |
ENSMUST00000028123.4
|
Gad2
|
glutamic acid decarboxylase 2 |
| chr3_-_154760978 | 0.78 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr5_-_23821523 | 0.77 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr11_+_102495189 | 0.76 |
ENSMUST00000057893.7
|
Fzd2
|
frizzled class receptor 2 |
| chr19_-_5610628 | 0.76 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr2_-_32976378 | 0.75 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr6_+_145879839 | 0.74 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr6_-_138398376 | 0.72 |
ENSMUST00000163065.8
|
Lmo3
|
LIM domain only 3 |
| chr11_+_66802807 | 0.72 |
ENSMUST00000123434.3
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr10_+_20024203 | 0.71 |
ENSMUST00000020173.16
|
Map7
|
microtubule-associated protein 7 |
| chr1_-_87322443 | 0.68 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr4_-_58499398 | 0.67 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr2_-_111843053 | 0.66 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr7_+_43430459 | 0.65 |
ENSMUST00000014058.11
|
Klk10
|
kallikrein related-peptidase 10 |
| chr5_-_5315968 | 0.65 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr2_+_9887427 | 0.64 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr5_-_116162415 | 0.62 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr3_+_62327089 | 0.61 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr11_-_20282684 | 0.61 |
ENSMUST00000004634.7
ENSMUST00000109594.8 |
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr15_+_28203872 | 0.60 |
ENSMUST00000067048.8
|
Dnah5
|
dynein, axonemal, heavy chain 5 |
| chr4_+_108576846 | 0.59 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr11_-_77079794 | 0.59 |
ENSMUST00000108400.8
|
Efcab5
|
EF-hand calcium binding domain 5 |
| chr1_+_172383499 | 0.58 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chrX_+_55500170 | 0.58 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr2_-_79287095 | 0.56 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr12_-_25147139 | 0.55 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr16_-_57575070 | 0.55 |
ENSMUST00000089332.5
|
Col8a1
|
collagen, type VIII, alpha 1 |
| chr6_+_136528155 | 0.55 |
ENSMUST00000186742.2
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr15_+_65682066 | 0.53 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr13_+_42834039 | 0.52 |
ENSMUST00000128646.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr11_-_97591150 | 0.52 |
ENSMUST00000018681.14
|
Pcgf2
|
polycomb group ring finger 2 |
| chr18_+_37952556 | 0.52 |
ENSMUST00000055935.11
|
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr11_+_95915366 | 0.50 |
ENSMUST00000103157.10
|
Gip
|
gastric inhibitory polypeptide |
| chr10_+_73782857 | 0.50 |
ENSMUST00000191709.6
ENSMUST00000193739.6 ENSMUST00000195531.6 |
Pcdh15
|
protocadherin 15 |
| chr16_-_37205277 | 0.49 |
ENSMUST00000114787.8
ENSMUST00000114782.8 ENSMUST00000114775.8 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr9_-_79885063 | 0.49 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chr9_-_96513529 | 0.49 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr3_+_55369149 | 0.49 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr9_+_61279640 | 0.49 |
ENSMUST00000178113.8
ENSMUST00000159386.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr1_+_177557380 | 0.48 |
ENSMUST00000016106.6
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr13_-_78345932 | 0.48 |
ENSMUST00000127137.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr15_-_64794139 | 0.48 |
ENSMUST00000023007.7
ENSMUST00000228014.2 |
Adcy8
|
adenylate cyclase 8 |
| chr2_+_152596075 | 0.48 |
ENSMUST00000010020.12
|
Cox4i2
|
cytochrome c oxidase subunit 4I2 |
| chr5_+_13448833 | 0.48 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr11_+_16207705 | 0.48 |
ENSMUST00000109645.9
ENSMUST00000109647.3 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr5_-_137530214 | 0.47 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr10_+_18345706 | 0.47 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr5_+_27466914 | 0.47 |
ENSMUST00000101471.4
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr15_-_8740218 | 0.46 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr18_+_37952596 | 0.46 |
ENSMUST00000193890.2
ENSMUST00000193941.2 |
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr4_-_58206596 | 0.46 |
ENSMUST00000042850.9
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr5_+_115573055 | 0.46 |
ENSMUST00000136586.6
|
Msi1
|
musashi RNA-binding protein 1 |
| chr5_-_131645437 | 0.45 |
ENSMUST00000161804.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr5_+_138185747 | 0.44 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr9_-_79884920 | 0.44 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1 |
| chr8_-_3674993 | 0.43 |
ENSMUST00000142431.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr11_-_33942981 | 0.43 |
ENSMUST00000238903.2
|
Kcnip1
|
Kv channel-interacting protein 1 |
| chr18_-_24736848 | 0.43 |
ENSMUST00000070726.10
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr1_+_11063678 | 0.43 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr5_-_90788460 | 0.42 |
ENSMUST00000202704.4
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr10_-_43934774 | 0.41 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr17_+_93506435 | 0.41 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr6_-_128252540 | 0.40 |
ENSMUST00000130454.8
|
Tead4
|
TEA domain family member 4 |
| chr3_-_92031247 | 0.40 |
ENSMUST00000070284.4
|
Prr9
|
proline rich 9 |
| chr8_+_24159669 | 0.40 |
ENSMUST00000042352.11
ENSMUST00000207301.2 |
Zmat4
|
zinc finger, matrin type 4 |
| chr6_-_102441628 | 0.39 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr18_-_24736521 | 0.39 |
ENSMUST00000154205.2
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chrM_+_7006 | 0.39 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr4_+_19818718 | 0.38 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr10_+_29074950 | 0.38 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr2_+_124978518 | 0.38 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr1_+_100025486 | 0.38 |
ENSMUST00000188735.2
|
Cntnap5b
|
contactin associated protein-like 5B |
| chr8_+_57774010 | 0.38 |
ENSMUST00000040104.5
|
Hand2
|
heart and neural crest derivatives expressed 2 |
| chr18_+_37898633 | 0.36 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr2_+_124978612 | 0.36 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr2_+_51928017 | 0.35 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr7_+_143792455 | 0.35 |
ENSMUST00000239495.2
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr15_-_8739893 | 0.35 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr6_-_23650205 | 0.34 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr13_+_75855695 | 0.33 |
ENSMUST00000222194.2
ENSMUST00000223535.2 ENSMUST00000222853.2 |
Ell2
|
elongation factor for RNA polymerase II 2 |
| chr6_-_23650297 | 0.33 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr6_+_8948608 | 0.33 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr3_+_10153163 | 0.33 |
ENSMUST00000061419.9
|
Gm9833
|
predicted gene 9833 |
| chr14_-_109151590 | 0.33 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr17_+_93506590 | 0.32 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr19_+_44980565 | 0.31 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr2_+_177969456 | 0.31 |
ENSMUST00000133267.3
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr11_-_99134885 | 0.30 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr2_-_17465410 | 0.29 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr14_+_32043944 | 0.29 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr13_+_55547498 | 0.27 |
ENSMUST00000057167.9
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr13_-_53627110 | 0.27 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr17_+_3447465 | 0.27 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr8_+_46111778 | 0.26 |
ENSMUST00000143820.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_-_77533596 | 0.26 |
ENSMUST00000171063.8
|
Zfp385b
|
zinc finger protein 385B |
| chr17_+_35150229 | 0.26 |
ENSMUST00000007253.6
|
Neu1
|
neuraminidase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 1.0 | 6.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.0 | 3.1 | GO:0061033 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.0 | 3.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.9 | 2.8 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.9 | 2.7 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.9 | 5.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.8 | 6.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.7 | 3.4 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.7 | 10.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.7 | 4.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.5 | 3.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 1.5 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.5 | 5.9 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.4 | 2.5 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.4 | 1.5 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.3 | 2.1 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.3 | 2.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 0.9 | GO:0060618 | nipple development(GO:0060618) |
| 0.3 | 1.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 1.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 5.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 1.0 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.3 | 0.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 0.8 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 2.3 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.2 | 2.4 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 | 0.7 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.2 | 3.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 1.7 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.2 | 0.6 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.2 | 0.8 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.2 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 0.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 0.7 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 6.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 4.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.5 | GO:0070346 | white fat cell proliferation(GO:0070343) positive regulation of fat cell proliferation(GO:0070346) regulation of white fat cell proliferation(GO:0070350) |
| 0.1 | 4.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 4.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 1.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 1.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.3 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.5 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.1 | 0.2 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 4.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.2 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 0.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 4.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 1.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 0.6 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.6 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.2 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.8 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.8 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 5.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 1.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 1.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 3.0 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.4 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.1 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 3.5 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 1.7 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 3.9 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 3.0 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.5 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.5 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 0.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 1.4 | 4.2 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.6 | 2.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.5 | 10.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 13.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 9.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 4.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.7 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 3.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 4.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 3.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 3.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 4.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 4.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 1.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 3.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 4.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 14.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 3.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 1.0 | 6.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.5 | 4.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.5 | 5.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 3.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.4 | 4.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 1.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 2.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.3 | 2.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 10.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 5.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 2.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 3.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 3.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 4.8 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 1.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 2.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 4.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 2.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 3.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.8 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 3.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.6 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 1.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.3 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 8.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 2.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 6.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 9.3 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 3.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 6.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 18.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 2.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 4.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 4.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 5.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 3.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 5.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 3.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 4.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 4.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 4.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.9 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 2.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 3.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 3.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |