Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
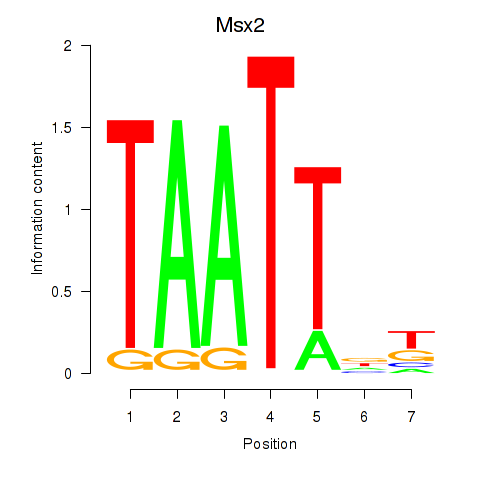
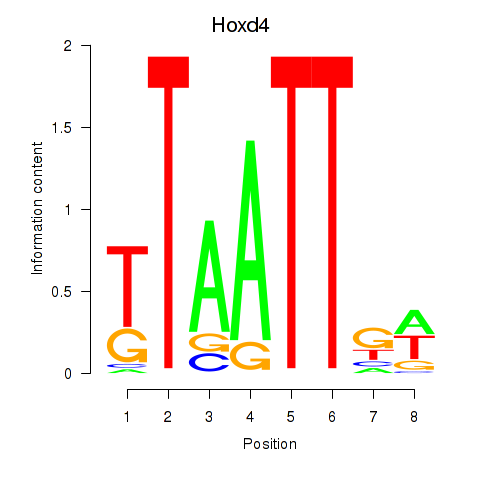
Results for Msx2_Hoxd4
Z-value: 0.65
Transcription factors associated with Msx2_Hoxd4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx2
|
ENSMUSG00000021469.10 | Msx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx2 | mm39_v1_chr13_-_53627110_53627110 | 0.12 | 3.1e-01 | Click! |
Activity profile of Msx2_Hoxd4 motif
Sorted Z-values of Msx2_Hoxd4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx2_Hoxd4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_6673167 | 6.33 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr11_+_67061908 | 4.94 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr11_+_67061837 | 4.77 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr1_-_144427302 | 2.90 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr3_-_14843512 | 2.84 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr3_-_75177378 | 2.71 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr5_-_62923463 | 2.64 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr6_+_68233361 | 2.22 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr4_-_96673423 | 2.17 |
ENSMUST00000107071.2
|
Gm12695
|
predicted gene 12695 |
| chr2_+_4022537 | 2.15 |
ENSMUST00000177457.8
|
Frmd4a
|
FERM domain containing 4A |
| chrX_+_158491589 | 2.06 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr18_+_4920513 | 2.05 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr3_+_40755211 | 1.88 |
ENSMUST00000204473.2
|
Plk4
|
polo like kinase 4 |
| chr13_+_49761506 | 1.86 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr13_+_49697919 | 1.80 |
ENSMUST00000177948.2
ENSMUST00000021820.14 |
Aspn
|
asporin |
| chr14_+_79753055 | 1.76 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr1_-_149836974 | 1.76 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr19_-_46033353 | 1.72 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr1_-_165535654 | 1.59 |
ENSMUST00000097474.9
|
Rcsd1
|
RCSD domain containing 1 |
| chr7_-_45570828 | 1.59 |
ENSMUST00000038876.13
|
Emp3
|
epithelial membrane protein 3 |
| chrX_+_55500170 | 1.55 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr1_+_45834645 | 1.54 |
ENSMUST00000147308.2
|
Wdr75
|
WD repeat domain 75 |
| chr2_-_84255602 | 1.51 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr6_-_136918885 | 1.49 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr7_-_45570254 | 1.46 |
ENSMUST00000164119.3
|
Emp3
|
epithelial membrane protein 3 |
| chr6_-_69584812 | 1.38 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr7_-_45570538 | 1.37 |
ENSMUST00000210297.2
|
Emp3
|
epithelial membrane protein 3 |
| chr6_+_149226891 | 1.36 |
ENSMUST00000189837.2
|
Resf1
|
retroelement silencing factor 1 |
| chrX_+_158086253 | 1.35 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr18_-_43610829 | 1.35 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr10_-_107321938 | 1.33 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr6_-_124756645 | 1.33 |
ENSMUST00000147669.2
ENSMUST00000128697.8 ENSMUST00000032218.10 ENSMUST00000112475.9 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr7_-_45570674 | 1.32 |
ENSMUST00000210939.2
|
Emp3
|
epithelial membrane protein 3 |
| chr3_-_49711706 | 1.29 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr2_-_72817060 | 1.28 |
ENSMUST00000112062.2
|
Gm11084
|
predicted gene 11084 |
| chr15_-_101422054 | 1.26 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr2_-_45002902 | 1.21 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr3_-_49711765 | 1.20 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr6_-_129449739 | 1.20 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr5_-_23821523 | 1.18 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr7_-_73191484 | 1.15 |
ENSMUST00000197642.2
ENSMUST00000026895.14 ENSMUST00000169922.9 |
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr13_+_19398273 | 1.11 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chr5_+_17779273 | 1.06 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr18_+_44237577 | 1.06 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr6_-_87510200 | 1.06 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr9_-_114219685 | 1.05 |
ENSMUST00000084881.5
|
Crtap
|
cartilage associated protein |
| chr3_+_32490300 | 1.04 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr10_-_63039709 | 0.99 |
ENSMUST00000095580.3
|
Mypn
|
myopalladin |
| chrX_+_132751729 | 0.95 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr6_-_129600798 | 0.94 |
ENSMUST00000095412.10
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr6_-_129600812 | 0.94 |
ENSMUST00000168919.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chrX_+_135145813 | 0.93 |
ENSMUST00000048687.11
|
Tceal9
|
transcription elongation factor A like 9 |
| chr7_-_120673761 | 0.93 |
ENSMUST00000047194.4
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr2_-_163259012 | 0.91 |
ENSMUST00000127038.2
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr4_-_3872105 | 0.91 |
ENSMUST00000105158.2
|
Mos
|
Moloney sarcoma oncogene |
| chr12_-_76224025 | 0.86 |
ENSMUST00000101291.11
ENSMUST00000218621.2 ENSMUST00000076634.5 |
Esr2
|
estrogen receptor 2 (beta) |
| chr9_-_107482494 | 0.83 |
ENSMUST00000102529.10
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr7_-_115459082 | 0.83 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr3_-_68911835 | 0.82 |
ENSMUST00000107812.8
|
Ift80
|
intraflagellar transport 80 |
| chr13_+_51562675 | 0.81 |
ENSMUST00000087978.5
|
S1pr3
|
sphingosine-1-phosphate receptor 3 |
| chr2_+_74522258 | 0.80 |
ENSMUST00000061745.5
|
Hoxd10
|
homeobox D10 |
| chr8_+_82582953 | 0.79 |
ENSMUST00000109851.3
|
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr3_-_146200870 | 0.78 |
ENSMUST00000093951.3
|
Spata1
|
spermatogenesis associated 1 |
| chr1_-_156766351 | 0.78 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr3_-_68911807 | 0.78 |
ENSMUST00000154741.8
ENSMUST00000148031.2 |
Ift80
|
intraflagellar transport 80 |
| chr2_-_73284262 | 0.77 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr16_+_14523696 | 0.77 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr19_+_45433899 | 0.75 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr4_-_82768958 | 0.74 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr11_+_98689479 | 0.73 |
ENSMUST00000037930.13
|
Msl1
|
male specific lethal 1 |
| chr7_+_140181182 | 0.71 |
ENSMUST00000214180.2
ENSMUST00000211771.2 |
Olfr46
|
olfactory receptor 46 |
| chrX_+_100683662 | 0.71 |
ENSMUST00000119299.8
ENSMUST00000044475.5 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) |
| chr9_-_107482462 | 0.68 |
ENSMUST00000194433.6
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr2_-_45000389 | 0.68 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr18_+_34973605 | 0.67 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr2_-_59955995 | 0.67 |
ENSMUST00000112550.8
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr8_+_31601837 | 0.66 |
ENSMUST00000046941.8
ENSMUST00000217278.2 |
Rnf122
|
ring finger protein 122 |
| chr7_-_113716996 | 0.66 |
ENSMUST00000069449.7
|
Rras2
|
related RAS viral (r-ras) oncogene 2 |
| chr12_-_113896002 | 0.65 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr9_-_107648144 | 0.65 |
ENSMUST00000183248.3
ENSMUST00000182022.8 ENSMUST00000035199.13 ENSMUST00000182659.8 |
Rbm5
|
RNA binding motif protein 5 |
| chr10_+_101994841 | 0.64 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr6_-_23650297 | 0.61 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr7_+_89814713 | 0.60 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_-_156766381 | 0.60 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr6_+_29361408 | 0.57 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chr6_+_124281607 | 0.57 |
ENSMUST00000032234.5
ENSMUST00000112541.8 |
Cd163
|
CD163 antigen |
| chrX_+_149330371 | 0.57 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr3_+_7494108 | 0.57 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr3_+_84859453 | 0.57 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr17_+_46471950 | 0.56 |
ENSMUST00000024748.14
ENSMUST00000172170.8 |
Gtpbp2
|
GTP binding protein 2 |
| chr11_-_79421397 | 0.56 |
ENSMUST00000103236.4
ENSMUST00000170799.8 ENSMUST00000170422.4 |
Evi2a
Evi2
|
ecotropic viral integration site 2a ecotropic viral integration site 2 |
| chr13_-_103042554 | 0.55 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr18_+_44237474 | 0.54 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr12_-_113733922 | 0.52 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr2_-_152422220 | 0.50 |
ENSMUST00000053180.4
|
Defb19
|
defensin beta 19 |
| chr10_+_5543769 | 0.50 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr13_+_21546990 | 0.50 |
ENSMUST00000225545.2
ENSMUST00000053293.14 |
Zscan12
|
zinc finger and SCAN domain containing 12 |
| chr1_-_80255156 | 0.49 |
ENSMUST00000168372.2
|
Cul3
|
cullin 3 |
| chr8_-_26609153 | 0.48 |
ENSMUST00000037182.14
|
Hook3
|
hook microtubule tethering protein 3 |
| chr3_-_144275897 | 0.48 |
ENSMUST00000043325.9
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr4_+_122730027 | 0.48 |
ENSMUST00000030412.11
ENSMUST00000121870.8 ENSMUST00000097902.5 |
Ppt1
|
palmitoyl-protein thioesterase 1 |
| chr5_-_121641461 | 0.48 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr13_-_21330037 | 0.48 |
ENSMUST00000216039.3
|
Olfr1368
|
olfactory receptor 1368 |
| chr18_+_9707595 | 0.47 |
ENSMUST00000234965.2
|
Colec12
|
collectin sub-family member 12 |
| chr3_-_75072319 | 0.47 |
ENSMUST00000124618.2
|
Zbbx
|
zinc finger, B-box domain containing |
| chr3_+_137770813 | 0.46 |
ENSMUST00000163080.3
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr1_+_165616315 | 0.45 |
ENSMUST00000161559.3
|
Cd247
|
CD247 antigen |
| chr2_+_20742115 | 0.45 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr13_-_103042294 | 0.44 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_-_89270554 | 0.44 |
ENSMUST00000220071.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr6_-_13871475 | 0.43 |
ENSMUST00000139231.2
|
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr2_+_89642395 | 0.43 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr14_-_36641270 | 0.43 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_+_53100796 | 0.42 |
ENSMUST00000027269.7
ENSMUST00000191197.2 |
Mstn
|
myostatin |
| chr4_+_109092610 | 0.42 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr1_-_155293141 | 0.41 |
ENSMUST00000111775.8
ENSMUST00000111774.2 |
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr9_-_56151334 | 0.41 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr17_-_37523969 | 0.41 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr16_+_27126239 | 0.41 |
ENSMUST00000066852.9
|
Ostn
|
osteocrin |
| chr19_+_26727111 | 0.41 |
ENSMUST00000175842.4
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_+_59197746 | 0.40 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr11_-_73742280 | 0.40 |
ENSMUST00000213365.2
|
Olfr393
|
olfactory receptor 393 |
| chr10_+_128139191 | 0.40 |
ENSMUST00000005825.8
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr18_+_7905440 | 0.40 |
ENSMUST00000170854.2
|
Wac
|
WW domain containing adaptor with coiled-coil |
| chr19_-_24178000 | 0.39 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr14_-_30973164 | 0.37 |
ENSMUST00000226565.2
ENSMUST00000022459.5 |
Phf7
|
PHD finger protein 7 |
| chr7_-_84328553 | 0.37 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr13_+_76115824 | 0.37 |
ENSMUST00000022081.3
|
Spata9
|
spermatogenesis associated 9 |
| chr14_+_54183465 | 0.37 |
ENSMUST00000197130.5
ENSMUST00000103677.3 |
Trdv2-1
|
T cell receptor delta variable 2-1 |
| chr6_-_23650205 | 0.37 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr6_-_57877975 | 0.36 |
ENSMUST00000228322.2
|
Vmn1r22
|
vomeronasal 1 receptor 22 |
| chr2_+_83554770 | 0.36 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V |
| chr14_+_53562089 | 0.35 |
ENSMUST00000178100.3
|
Trav7n-6
|
T cell receptor alpha variable 7N-6 |
| chr1_-_132318039 | 0.34 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr14_-_86986541 | 0.34 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr18_-_38102799 | 0.34 |
ENSMUST00000166148.8
ENSMUST00000163131.8 ENSMUST00000043437.14 |
Fchsd1
|
FCH and double SH3 domains 1 |
| chr17_-_29226700 | 0.34 |
ENSMUST00000233441.2
|
Stk38
|
serine/threonine kinase 38 |
| chr1_+_171668122 | 0.34 |
ENSMUST00000135386.2
|
Cd84
|
CD84 antigen |
| chr4_-_43710231 | 0.34 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr2_+_85809620 | 0.34 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr13_-_43634695 | 0.34 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr14_-_50586329 | 0.33 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr5_-_53864874 | 0.33 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr5_-_151351628 | 0.33 |
ENSMUST00000202365.2
ENSMUST00000186838.2 |
Gm42906
D730045B01Rik
|
predicted gene 42906 RIKEN cDNA D730045B01 gene |
| chr9_+_30853837 | 0.33 |
ENSMUST00000068135.13
|
Adamts8
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 8 |
| chr9_+_37400317 | 0.33 |
ENSMUST00000239459.2
|
Msantd2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr2_-_104324035 | 0.33 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr15_+_41694317 | 0.32 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr1_+_33947250 | 0.32 |
ENSMUST00000183034.5
|
Dst
|
dystonin |
| chr15_-_3333003 | 0.31 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr2_+_57887896 | 0.31 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr11_-_87716849 | 0.29 |
ENSMUST00000103177.10
|
Lpo
|
lactoperoxidase |
| chr10_-_14421368 | 0.29 |
ENSMUST00000208429.3
|
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr10_+_127256736 | 0.29 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2 |
| chr6_+_83142902 | 0.29 |
ENSMUST00000077407.12
ENSMUST00000113913.8 ENSMUST00000130212.8 |
Dctn1
|
dynactin 1 |
| chr14_+_53599724 | 0.28 |
ENSMUST00000196105.2
|
Trav13n-4
|
T cell receptor alpha variable 13N-4 |
| chr13_+_118851214 | 0.28 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr18_+_37453427 | 0.28 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr10_+_127257077 | 0.27 |
ENSMUST00000168780.8
|
R3hdm2
|
R3H domain containing 2 |
| chr18_-_3309858 | 0.26 |
ENSMUST00000144496.8
ENSMUST00000154715.8 |
Crem
|
cAMP responsive element modulator |
| chr6_-_40906665 | 0.26 |
ENSMUST00000136499.2
|
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
| chr11_-_99482165 | 0.25 |
ENSMUST00000104930.2
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr11_-_66059330 | 0.25 |
ENSMUST00000080665.10
|
Dnah9
|
dynein, axonemal, heavy chain 9 |
| chr5_-_123127346 | 0.24 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr11_-_99265721 | 0.24 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr15_-_36165017 | 0.24 |
ENSMUST00000058643.4
|
Fbxo43
|
F-box protein 43 |
| chr14_-_56137697 | 0.24 |
ENSMUST00000111325.5
|
Sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr1_-_14380418 | 0.23 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chrX_+_21350783 | 0.23 |
ENSMUST00000089188.9
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr10_+_127256993 | 0.23 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chr2_-_32976378 | 0.23 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr19_-_55229668 | 0.22 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr7_+_122723365 | 0.21 |
ENSMUST00000205514.2
ENSMUST00000094053.7 |
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr6_-_39702381 | 0.21 |
ENSMUST00000002487.15
|
Braf
|
Braf transforming gene |
| chr10_-_129107354 | 0.20 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr5_-_72325482 | 0.20 |
ENSMUST00000196241.2
ENSMUST00000013693.11 |
Commd8
|
COMM domain containing 8 |
| chr15_+_38869415 | 0.20 |
ENSMUST00000179165.9
|
Fzd6
|
frizzled class receptor 6 |
| chr6_+_15196950 | 0.19 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr3_+_144283355 | 0.19 |
ENSMUST00000151086.3
|
Selenof
|
selenoprotein F |
| chr6_+_29853745 | 0.19 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chrX_+_105626790 | 0.19 |
ENSMUST00000101296.9
ENSMUST00000101297.4 |
Gm5127
|
predicted gene 5127 |
| chr12_+_108145802 | 0.19 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr9_-_96513529 | 0.19 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr6_+_24733239 | 0.19 |
ENSMUST00000031690.6
|
Hyal6
|
hyaluronoglucosaminidase 6 |
| chr15_+_58004793 | 0.18 |
ENSMUST00000227142.2
ENSMUST00000226955.2 |
Wdyhv1
|
WDYHV motif containing 1 |
| chr6_+_83985684 | 0.18 |
ENSMUST00000203803.3
ENSMUST00000204591.3 ENSMUST00000113823.8 ENSMUST00000153860.4 |
Dysf
|
dysferlin |
| chr1_+_179788037 | 0.18 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr5_+_107655487 | 0.17 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr1_+_60948149 | 0.17 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr14_+_8283087 | 0.17 |
ENSMUST00000206298.3
ENSMUST00000216079.2 |
Olfr720
|
olfactory receptor 720 |
| chr11_-_99494134 | 0.17 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr1_+_179788675 | 0.17 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr19_-_47680528 | 0.17 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr5_-_82271183 | 0.15 |
ENSMUST00000186079.2
ENSMUST00000185607.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr7_-_101494472 | 0.15 |
ENSMUST00000211566.2
ENSMUST00000094141.7 ENSMUST00000209329.2 |
Folr2
|
folate receptor 2 (fetal) |
| chr9_+_38788422 | 0.14 |
ENSMUST00000078289.3
|
Olfr926
|
olfactory receptor 926 |
| chr15_-_36164963 | 0.14 |
ENSMUST00000227793.2
|
Fbxo43
|
F-box protein 43 |
| chr11_-_99457456 | 0.14 |
ENSMUST00000055502.5
|
Krtap3-1
|
keratin associated protein 3-1 |
| chr1_+_128079543 | 0.13 |
ENSMUST00000189317.3
|
R3hdm1
|
R3H domain containing 1 |
| chrX_+_106836189 | 0.13 |
ENSMUST00000101292.9
|
Tent5d
|
terminal nucleotidyltransferase 5D |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.6 | 1.7 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.5 | 9.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 1.5 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.4 | 1.8 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 1.0 | GO:1901874 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.3 | 1.0 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.3 | 0.9 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.3 | 0.9 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.3 | 1.9 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.3 | 0.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.3 | 1.8 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.7 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 1.9 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.2 | 0.6 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.2 | 1.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 5.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.6 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 1.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 1.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 1.6 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.1 | 1.6 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.4 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.1 | 1.4 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 1.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.4 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.1 | 0.6 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.3 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.1 | 0.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.6 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.5 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.1 | 0.3 | GO:0061033 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.4 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.1 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.5 | GO:0071630 | trophectodermal cellular morphogenesis(GO:0001831) nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.1 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.2 | GO:0021993 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.2 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.1 | 6.3 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.4 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 3.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.8 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 1.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 2.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.9 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 1.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.9 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.5 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.8 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.4 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) phosphate ion transmembrane transport(GO:0035435) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.3 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.0 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.4 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 1.4 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.6 | GO:0006953 | acute-phase response(GO:0006953) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 1.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 1.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.6 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 1.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 2.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 1.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 4.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.5 | 1.5 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.3 | 0.9 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 2.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 0.7 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.8 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 1.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.6 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 1.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.5 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 2.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 1.0 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0070773 | protein-N-terminal glutamine amidohydrolase activity(GO:0070773) |
| 0.1 | 0.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 2.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 4.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.5 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 1.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.5 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 2.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 3.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 2.3 | GO:0003823 | antigen binding(GO:0003823) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 6.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 1.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 2.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 5.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 3.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |