Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
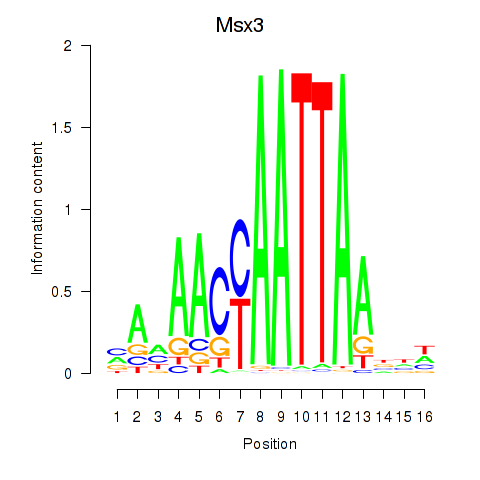
Results for Msx3
Z-value: 0.78
Transcription factors associated with Msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx3
|
ENSMUSG00000025469.11 | Msx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx3 | mm39_v1_chr7_-_139628991_139629007 | 0.19 | 1.2e-01 | Click! |
Activity profile of Msx3 motif
Sorted Z-values of Msx3 motif
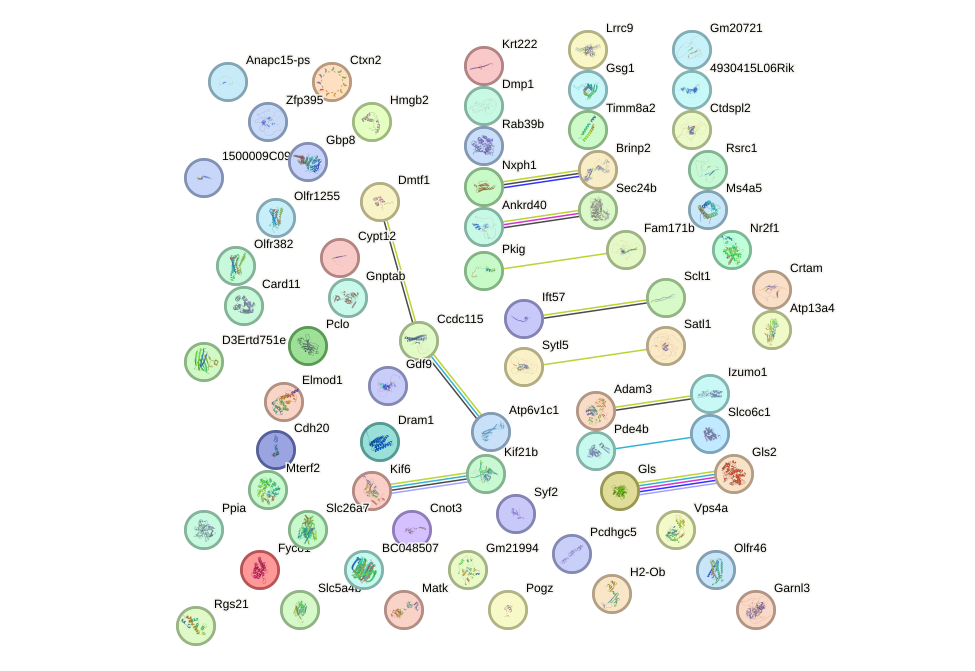
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_32977182 | 6.05 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr2_-_32976378 | 5.94 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr2_+_96148418 | 5.40 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr11_+_69909245 | 4.89 |
ENSMUST00000231415.2
ENSMUST00000108588.9 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr1_-_52230062 | 4.74 |
ENSMUST00000156887.8
ENSMUST00000129107.2 |
Gls
|
glutaminase |
| chr15_+_82136598 | 4.70 |
ENSMUST00000136948.3
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr10_+_81093110 | 4.56 |
ENSMUST00000117488.8
ENSMUST00000105328.10 ENSMUST00000121205.8 |
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr14_+_60615128 | 4.36 |
ENSMUST00000022561.9
|
Amer2
|
APC membrane recruitment 2 |
| chr18_+_37952556 | 4.12 |
ENSMUST00000055935.11
|
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr3_+_66893280 | 3.89 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr3_+_66892979 | 3.59 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr8_-_25215778 | 3.58 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr12_+_102094977 | 3.53 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr10_-_75946790 | 3.35 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr19_+_13208692 | 3.23 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr16_+_49519561 | 3.21 |
ENSMUST00000046777.11
ENSMUST00000142682.9 |
Ift57
|
intraflagellar transport 57 |
| chrX_-_74621828 | 3.14 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr5_+_14564932 | 3.12 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr3_+_66893031 | 3.10 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr7_+_45271229 | 2.97 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr11_-_99134885 | 2.90 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr4_+_102112189 | 2.89 |
ENSMUST00000106908.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr12_+_72488625 | 2.88 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr6_+_8948608 | 2.76 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr2_+_83642910 | 2.70 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr18_+_37952596 | 2.54 |
ENSMUST00000193890.2
ENSMUST00000193941.2 |
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr2_+_87696836 | 2.52 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr18_+_37827413 | 2.46 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr1_-_171854818 | 2.37 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr2_+_124978518 | 2.34 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr7_+_101545547 | 2.31 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr5_+_88731386 | 2.24 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr1_+_104696235 | 2.16 |
ENSMUST00000062528.9
|
Cdh20
|
cadherin 20 |
| chr8_+_107757847 | 2.06 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr15_+_38662158 | 2.02 |
ENSMUST00000022904.8
ENSMUST00000228820.2 |
Atp6v1c1
|
ATPase, H+ transporting, lysosomal V1 subunit C1 |
| chr16_-_29363671 | 2.02 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr5_+_88731366 | 1.99 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr2_+_124978612 | 1.94 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr17_+_34457868 | 1.88 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chrX_-_111315519 | 1.86 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr6_-_135231324 | 1.86 |
ENSMUST00000111911.9
ENSMUST00000111910.4 |
Gsg1
|
germ cell associated 1 |
| chr7_+_140181182 | 1.85 |
ENSMUST00000214180.2
ENSMUST00000211771.2 |
Olfr46
|
olfactory receptor 46 |
| chr7_+_143838191 | 1.80 |
ENSMUST00000097929.4
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr10_+_29074950 | 1.78 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chrX_-_142716200 | 1.74 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr3_+_18002574 | 1.74 |
ENSMUST00000029080.5
|
Cypt12
|
cysteine-rich perinuclear theca 12 |
| chr1_-_97055931 | 1.69 |
ENSMUST00000027569.14
|
Slco6c1
|
solute carrier organic anion transporter family, member 6c1 |
| chr13_-_78344492 | 1.69 |
ENSMUST00000125176.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr3_-_41696906 | 1.66 |
ENSMUST00000026866.15
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr19_-_11261177 | 1.61 |
ENSMUST00000186937.7
ENSMUST00000067673.13 |
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr1_-_158183894 | 1.60 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr5_+_104350475 | 1.52 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr9_-_53882530 | 1.51 |
ENSMUST00000048409.14
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr6_+_134617903 | 1.42 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chrX_+_106299484 | 1.39 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr12_-_114443071 | 1.37 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr13_+_68011442 | 1.36 |
ENSMUST00000078471.7
|
BC048507
|
cDNA sequence BC048507 |
| chr4_+_134658209 | 1.33 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr6_-_135231168 | 1.32 |
ENSMUST00000111909.8
|
Gsg1
|
germ cell associated 1 |
| chr10_-_84963841 | 1.28 |
ENSMUST00000214193.2
ENSMUST00000050813.4 ENSMUST00000217027.2 |
Mterf2
|
mitochondrial transcription termination factor 2 |
| chr4_+_102111936 | 1.28 |
ENSMUST00000106907.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_45007407 | 1.24 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr8_+_66964401 | 1.21 |
ENSMUST00000002025.5
ENSMUST00000183187.2 |
Tktl2
|
transketolase-like 2 |
| chr7_+_101546059 | 1.17 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chrX_+_152506577 | 1.14 |
ENSMUST00000140575.8
ENSMUST00000208373.2 ENSMUST00000185492.7 ENSMUST00000149514.8 |
Nbdy
|
negative regulator of P-body association |
| chr2_+_163500290 | 1.08 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr14_+_122272069 | 1.07 |
ENSMUST00000045976.7
|
Timm8a2
|
translocase of inner mitochondrial membrane 8A2 |
| chr4_-_43710231 | 1.03 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr17_+_30940015 | 1.00 |
ENSMUST00000236948.2
|
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chrX_-_88976458 | 0.96 |
ENSMUST00000088146.3
|
4930415L06Rik
|
RIKEN cDNA 4930415L06 gene |
| chr14_+_65612788 | 0.95 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr2_+_121786892 | 0.87 |
ENSMUST00000110578.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr2_-_174188505 | 0.86 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chr2_+_121787131 | 0.84 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chrX_+_9751861 | 0.83 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr17_-_24746911 | 0.81 |
ENSMUST00000176652.8
|
Traf7
|
TNF receptor-associated factor 7 |
| chr8_+_57964921 | 0.81 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr11_+_109434519 | 0.81 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr3_-_129834788 | 0.80 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr10_+_88215079 | 0.80 |
ENSMUST00000130301.8
ENSMUST00000020251.10 |
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr9_-_40895756 | 0.80 |
ENSMUST00000180872.9
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr5_-_140986312 | 0.78 |
ENSMUST00000085786.7
|
Card11
|
caspase recruitment domain family, member 11 |
| chr7_+_89814713 | 0.76 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr5_-_105198913 | 0.73 |
ENSMUST00000112718.5
|
Gbp8
|
guanylate-binding protein 8 |
| chr3_+_94745009 | 0.68 |
ENSMUST00000107266.8
ENSMUST00000042402.12 ENSMUST00000107269.2 |
Pogz
|
pogo transposable element with ZNF domain |
| chr16_+_11224481 | 0.66 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr2_+_87686206 | 0.65 |
ENSMUST00000217376.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr17_+_49922129 | 0.65 |
ENSMUST00000162854.2
|
Kif6
|
kinesin family member 6 |
| chr17_+_21603985 | 0.61 |
ENSMUST00000056107.11
ENSMUST00000162659.2 |
Zfp677
|
zinc finger protein 677 |
| chr11_+_6366259 | 0.57 |
ENSMUST00000213200.2
|
Ppia
|
peptidylprolyl isomerase A |
| chr2_-_111843053 | 0.57 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr11_+_53324126 | 0.56 |
ENSMUST00000018382.7
|
Gdf9
|
growth differentiation factor 9 |
| chr2_+_89642395 | 0.50 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr2_-_150097511 | 0.40 |
ENSMUST00000063463.6
|
Gm21994
|
predicted gene 21994 |
| chrX_-_142716085 | 0.38 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr13_+_22508759 | 0.36 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr7_+_3648264 | 0.35 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr10_+_88214989 | 0.34 |
ENSMUST00000127615.8
|
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr1_-_144427302 | 0.30 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr3_+_94744844 | 0.30 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain |
| chr11_-_73382303 | 0.30 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr17_-_24746804 | 0.21 |
ENSMUST00000176353.8
ENSMUST00000176237.8 |
Traf7
|
TNF receptor-associated factor 7 |
| chr3_+_41697046 | 0.20 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr2_-_87504008 | 0.20 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr13_+_19394484 | 0.19 |
ENSMUST00000200495.2
|
Trgj1
|
T cell receptor gamma joining 1 |
| chr4_-_14621497 | 0.19 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr9_+_38516398 | 0.18 |
ENSMUST00000217057.2
|
Olfr914
|
olfactory receptor 914 |
| chr11_+_94218810 | 0.18 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr8_+_57964956 | 0.16 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr10_-_88214913 | 0.15 |
ENSMUST00000138159.2
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr8_+_46080746 | 0.12 |
ENSMUST00000145458.9
ENSMUST00000134321.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr13_+_22268610 | 0.09 |
ENSMUST00000228243.2
ENSMUST00000226680.2 |
Vmn1r188
|
vomeronasal 1 receptor 188 |
| chr1_-_34478753 | 0.07 |
ENSMUST00000042493.10
|
Ccdc115
|
coiled-coil domain containing 115 |
| chr13_+_23191826 | 0.06 |
ENSMUST00000228758.2
ENSMUST00000228031.2 ENSMUST00000227573.2 |
Vmn1r213
|
vomeronasal 1 receptor 213 |
| chr13_-_23420117 | 0.05 |
ENSMUST00000235574.2
ENSMUST00000226672.3 |
Vmn1r222
|
vomeronasal 1 receptor 222 |
| chr1_+_177557380 | 0.03 |
ENSMUST00000016106.6
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr19_+_12257218 | 0.02 |
ENSMUST00000207186.4
ENSMUST00000207915.2 |
Olfr1434
|
olfactory receptor 1434 |
| chr3_+_82962823 | 0.01 |
ENSMUST00000150268.8
ENSMUST00000122128.2 |
Plrg1
|
pleiotropic regulator 1 |
| chr9_+_72714156 | 0.00 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.8 | 2.4 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.8 | 4.7 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.7 | 2.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.6 | 1.9 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.4 | 1.3 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.4 | 1.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.4 | 1.9 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.3 | 4.9 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.3 | 4.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 1.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.3 | 0.8 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.2 | 3.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.2 | 0.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 3.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 1.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 1.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 3.0 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 3.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 4.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.8 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 1.4 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 3.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 2.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.6 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 7.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 1.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 3.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 5.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.0 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.8 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 1.8 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 1.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 4.6 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 4.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 1.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 11.9 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 9.8 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.4 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.5 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.8 | 2.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.5 | 2.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 4.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 1.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.2 | 2.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 4.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 1.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 4.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 3.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 3.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 3.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 10.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 4.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 5.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.7 | 4.9 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.6 | 3.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.5 | 1.9 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.4 | 1.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.3 | 1.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.3 | 3.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 3.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 3.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 3.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 4.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 2.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 4.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 6.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 4.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 13.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 2.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 4.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 4.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |