Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Mtf1
Z-value: 0.86
Transcription factors associated with Mtf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mtf1
|
ENSMUSG00000028890.14 | Mtf1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mtf1 | mm39_v1_chr4_+_124696336_124696381 | 0.36 | 1.9e-03 | Click! |
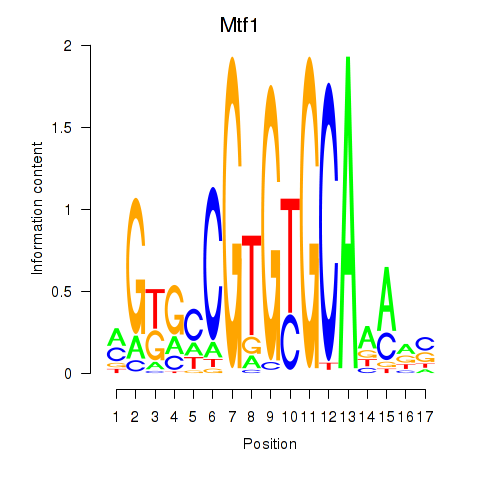
Activity profile of Mtf1 motif
Sorted Z-values of Mtf1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Mtf1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_94899292 | 12.89 |
ENSMUST00000034214.8
ENSMUST00000212806.2 |
Mt2
|
metallothionein 2 |
| chr8_+_94905710 | 5.07 |
ENSMUST00000034215.8
ENSMUST00000212291.2 ENSMUST00000211807.2 |
Mt1
|
metallothionein 1 |
| chr1_+_66426127 | 4.80 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr10_-_80837173 | 4.72 |
ENSMUST00000099462.8
ENSMUST00000118233.8 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chrX_-_8072714 | 3.96 |
ENSMUST00000089403.10
ENSMUST00000077595.12 ENSMUST00000089402.10 ENSMUST00000082320.12 |
Porcn
|
porcupine O-acyltransferase |
| chr9_+_98372575 | 3.93 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr4_+_134070492 | 3.77 |
ENSMUST00000105873.8
ENSMUST00000105874.9 |
Slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr3_+_107803225 | 3.65 |
ENSMUST00000172247.8
ENSMUST00000167387.8 |
Gstm5
|
glutathione S-transferase, mu 5 |
| chr2_+_28082943 | 3.53 |
ENSMUST00000113920.8
|
Olfm1
|
olfactomedin 1 |
| chr9_-_14292269 | 3.50 |
ENSMUST00000214236.2
|
Endod1
|
endonuclease domain containing 1 |
| chr11_+_63023893 | 3.47 |
ENSMUST00000108700.2
|
Pmp22
|
peripheral myelin protein 22 |
| chr3_+_107803137 | 3.40 |
ENSMUST00000004134.11
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr7_-_46782448 | 3.39 |
ENSMUST00000033142.13
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr10_+_20828446 | 3.22 |
ENSMUST00000105525.12
|
Ahi1
|
Abelson helper integration site 1 |
| chr3_+_107803563 | 3.21 |
ENSMUST00000169365.2
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr11_+_63023395 | 3.17 |
ENSMUST00000108701.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr12_+_111725357 | 3.15 |
ENSMUST00000118471.8
ENSMUST00000122300.8 |
Klc1
|
kinesin light chain 1 |
| chr4_+_134070777 | 2.90 |
ENSMUST00000105872.8
|
Slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr6_-_126717590 | 2.84 |
ENSMUST00000185333.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr12_+_111725282 | 2.79 |
ENSMUST00000239017.2
ENSMUST00000084941.12 |
Klc1
|
kinesin light chain 1 |
| chr5_+_57875309 | 2.71 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr9_+_34400117 | 2.59 |
ENSMUST00000188933.7
ENSMUST00000187182.7 |
Kirrel3
|
kirre like nephrin family adhesion molecule 3 |
| chr9_-_112016966 | 2.45 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr7_+_43228999 | 2.44 |
ENSMUST00000058104.8
ENSMUST00000205769.2 |
Zfp719
|
zinc finger protein 719 |
| chr1_+_135656885 | 2.43 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr9_+_34400082 | 2.41 |
ENSMUST00000190549.7
ENSMUST00000045091.13 |
Kirrel3
|
kirre like nephrin family adhesion molecule 3 |
| chr18_+_11972277 | 2.38 |
ENSMUST00000171109.9
ENSMUST00000046948.10 |
Cables1
|
CDK5 and Abl enzyme substrate 1 |
| chr6_-_126717114 | 2.32 |
ENSMUST00000112242.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr1_-_37758863 | 2.31 |
ENSMUST00000160589.2
|
Cracdl
|
capping protein inhibiting regulator of actin like |
| chr15_+_27466732 | 2.29 |
ENSMUST00000022875.7
|
Ank
|
progressive ankylosis |
| chr1_+_75376714 | 2.18 |
ENSMUST00000113589.8
|
Speg
|
SPEG complex locus |
| chr19_-_4665668 | 1.87 |
ENSMUST00000113822.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr7_+_140500808 | 1.85 |
ENSMUST00000106045.8
ENSMUST00000183845.8 |
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr8_+_124100492 | 1.84 |
ENSMUST00000212571.2
ENSMUST00000212470.2 ENSMUST00000108840.4 ENSMUST00000057934.10 |
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr19_-_6910195 | 1.84 |
ENSMUST00000236443.2
|
Kcnk4
|
potassium channel, subfamily K, member 4 |
| chr14_-_63781381 | 1.82 |
ENSMUST00000058679.7
|
Mtmr9
|
myotubularin related protein 9 |
| chr10_-_117074501 | 1.81 |
ENSMUST00000159193.8
ENSMUST00000020392.5 |
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
| chr7_+_140500848 | 1.77 |
ENSMUST00000184560.2
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr1_+_127132712 | 1.75 |
ENSMUST00000038361.11
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr1_+_93948173 | 1.65 |
ENSMUST00000190212.7
ENSMUST00000050890.8 |
Neu4
|
sialidase 4 |
| chr8_-_71229293 | 1.65 |
ENSMUST00000034296.15
|
Pik3r2
|
phosphoinositide-3-kinase regulatory subunit 2 |
| chr19_-_4665509 | 1.64 |
ENSMUST00000053597.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr7_-_15656285 | 1.58 |
ENSMUST00000044355.11
|
Selenow
|
selenoprotein W |
| chr9_+_34400016 | 1.49 |
ENSMUST00000187625.7
|
Kirrel3
|
kirre like nephrin family adhesion molecule 3 |
| chr15_-_58078274 | 1.46 |
ENSMUST00000022986.8
|
Fbxo32
|
F-box protein 32 |
| chr15_+_81686622 | 1.43 |
ENSMUST00000109553.10
|
Tef
|
thyrotroph embryonic factor |
| chr9_-_66032134 | 1.40 |
ENSMUST00000034946.15
|
Snx1
|
sorting nexin 1 |
| chr4_-_43040278 | 1.38 |
ENSMUST00000107958.8
ENSMUST00000107959.8 ENSMUST00000152846.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr15_-_93417380 | 1.11 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr1_+_128031055 | 1.10 |
ENSMUST00000188381.7
ENSMUST00000187900.7 ENSMUST00000036288.11 |
R3hdm1
|
R3H domain containing 1 |
| chr11_+_50022167 | 1.00 |
ENSMUST00000093138.13
ENSMUST00000101270.5 |
Tbc1d9b
|
TBC1 domain family, member 9B |
| chr10_+_29023201 | 0.99 |
ENSMUST00000213243.2
|
Soga3
|
SOGA family member 3 |
| chr16_-_91394522 | 0.95 |
ENSMUST00000023686.15
|
Tmem50b
|
transmembrane protein 50B |
| chr4_+_152171286 | 0.95 |
ENSMUST00000118648.8
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr11_+_58845502 | 0.94 |
ENSMUST00000108817.5
ENSMUST00000047697.12 |
H2aw
Trim17
|
H2A.W histone tripartite motif-containing 17 |
| chr8_+_106937625 | 0.83 |
ENSMUST00000109297.8
|
Prmt7
|
protein arginine N-methyltransferase 7 |
| chr8_+_124100588 | 0.81 |
ENSMUST00000211932.2
ENSMUST00000212569.2 |
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr11_-_32172233 | 0.78 |
ENSMUST00000150381.2
ENSMUST00000144902.2 ENSMUST00000020524.15 |
Rhbdf1
|
rhomboid 5 homolog 1 |
| chr8_+_106937568 | 0.76 |
ENSMUST00000071592.12
|
Prmt7
|
protein arginine N-methyltransferase 7 |
| chr17_+_47999916 | 0.76 |
ENSMUST00000156118.8
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr10_-_7162196 | 0.72 |
ENSMUST00000015346.12
|
Cnksr3
|
Cnksr family member 3 |
| chr16_-_18165876 | 0.66 |
ENSMUST00000125287.9
|
Tango2
|
transport and golgi organization 2 |
| chr18_-_77652820 | 0.64 |
ENSMUST00000026494.14
ENSMUST00000182024.2 |
Rnf165
|
ring finger protein 165 |
| chr13_-_24118139 | 0.64 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr5_-_34794451 | 0.60 |
ENSMUST00000124668.2
ENSMUST00000001109.11 ENSMUST00000155577.8 ENSMUST00000114329.8 |
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr5_+_122788469 | 0.60 |
ENSMUST00000199371.2
|
P2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr3_+_54662995 | 0.59 |
ENSMUST00000029371.3
|
Smad9
|
SMAD family member 9 |
| chr5_+_31026967 | 0.59 |
ENSMUST00000114716.4
|
Tmem214
|
transmembrane protein 214 |
| chr15_+_100366966 | 0.59 |
ENSMUST00000229648.2
|
Letmd1
|
LETM1 domain containing 1 |
| chr14_+_30673334 | 0.58 |
ENSMUST00000226551.2
ENSMUST00000228328.2 |
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr16_-_22676264 | 0.48 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr5_-_34794185 | 0.47 |
ENSMUST00000149657.5
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr15_+_100366886 | 0.45 |
ENSMUST00000037001.10
ENSMUST00000230294.2 |
Letmd1
|
LETM1 domain containing 1 |
| chr7_+_132212349 | 0.41 |
ENSMUST00000033241.6
ENSMUST00000106170.8 |
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr8_+_71358576 | 0.39 |
ENSMUST00000019405.4
ENSMUST00000212511.2 |
Map1s
|
microtubule-associated protein 1S |
| chr10_+_80972089 | 0.38 |
ENSMUST00000048128.15
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr2_-_91067312 | 0.38 |
ENSMUST00000028696.5
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr13_+_24118417 | 0.37 |
ENSMUST00000072391.2
|
H2ac1
|
H2A clustered histone 1 |
| chr14_+_59438658 | 0.37 |
ENSMUST00000173547.8
ENSMUST00000043227.13 ENSMUST00000022551.14 |
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr11_+_4845328 | 0.37 |
ENSMUST00000038237.8
|
Thoc5
|
THO complex 5 |
| chr14_+_59438879 | 0.36 |
ENSMUST00000140136.9
ENSMUST00000142326.2 |
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr11_+_29323618 | 0.36 |
ENSMUST00000040182.13
ENSMUST00000109477.2 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr2_-_119617985 | 0.34 |
ENSMUST00000110793.8
ENSMUST00000099529.9 ENSMUST00000048493.12 |
Rpap1
|
RNA polymerase II associated protein 1 |
| chr16_+_56942050 | 0.32 |
ENSMUST00000166897.3
|
Tomm70a
|
translocase of outer mitochondrial membrane 70A |
| chr5_-_34794546 | 0.30 |
ENSMUST00000114331.10
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr6_-_39095144 | 0.29 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr9_+_19716202 | 0.26 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr8_+_13209141 | 0.25 |
ENSMUST00000033824.8
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr7_-_42962487 | 0.21 |
ENSMUST00000135130.2
ENSMUST00000139061.2 |
Zfp715
|
zinc finger protein 715 |
| chr7_+_126447080 | 0.21 |
ENSMUST00000147257.2
ENSMUST00000139174.2 |
Doc2a
|
double C2, alpha |
| chr14_+_30673409 | 0.21 |
ENSMUST00000050171.10
|
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr11_+_4207557 | 0.15 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr14_+_30673435 | 0.14 |
ENSMUST00000226833.2
|
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr5_-_112503899 | 0.14 |
ENSMUST00000051117.3
|
Gm6583
|
predicted gene 6583 |
| chr4_-_126057263 | 0.14 |
ENSMUST00000097891.4
|
Sh3d21
|
SH3 domain containing 21 |
| chr4_+_134224308 | 0.12 |
ENSMUST00000095074.4
|
Paqr7
|
progestin and adipoQ receptor family member VII |
| chr11_+_4845314 | 0.10 |
ENSMUST00000101615.9
|
Thoc5
|
THO complex 5 |
| chr2_-_130506484 | 0.09 |
ENSMUST00000089559.11
|
Ddrgk1
|
DDRGK domain containing 1 |
| chr3_+_104688363 | 0.06 |
ENSMUST00000002298.7
|
Ppm1j
|
protein phosphatase 1J |
| chr2_-_91067212 | 0.05 |
ENSMUST00000111352.8
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr2_+_30485048 | 0.05 |
ENSMUST00000102853.4
|
Cstad
|
CSA-conditional, T cell activation-dependent protein |
| chr3_-_121325887 | 0.05 |
ENSMUST00000039197.9
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr7_-_119393182 | 0.01 |
ENSMUST00000106523.8
ENSMUST00000063902.14 ENSMUST00000150844.3 |
Eri2
|
exoribonuclease 2 |
| chr11_-_5848771 | 0.00 |
ENSMUST00000102921.4
|
Myl7
|
myosin, light polypeptide 7, regulatory |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 18.0 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 2.2 | 6.7 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 1.7 | 10.3 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 1.3 | 6.5 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 1.2 | 5.9 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 1.1 | 3.2 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.8 | 3.4 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.5 | 1.5 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.5 | 4.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.5 | 1.8 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.4 | 3.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.4 | 1.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.3 | 1.4 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.3 | 0.9 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.3 | 3.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 7.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.6 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 1.7 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 3.6 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.2 | 2.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 4.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 1.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.6 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.2 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.5 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 4.0 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 0.2 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 2.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 1.6 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.7 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.9 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 2.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 5.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.6 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 2.2 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 1.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.4 | 10.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 6.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 6.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 3.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 3.6 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 3.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 7.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.2 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.1 | 4.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 6.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 7.0 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 1.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 6.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.4 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.6 | 1.8 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.5 | 4.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 1.8 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.4 | 2.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.4 | 3.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 10.3 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.4 | 1.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.3 | 1.4 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.3 | 1.7 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 1.6 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 3.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 5.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 1.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 6.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 1.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 3.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 5.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 6.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.6 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 2.4 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 11.7 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 6.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 4.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 4.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.8 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 10.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 8.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 5.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 4.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 5.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.9 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |