Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Myb
Z-value: 3.80
Transcription factors associated with Myb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myb
|
ENSMUSG00000019982.16 | Myb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myb | mm39_v1_chr10_-_21036792_21036810 | 0.41 | 2.9e-04 | Click! |
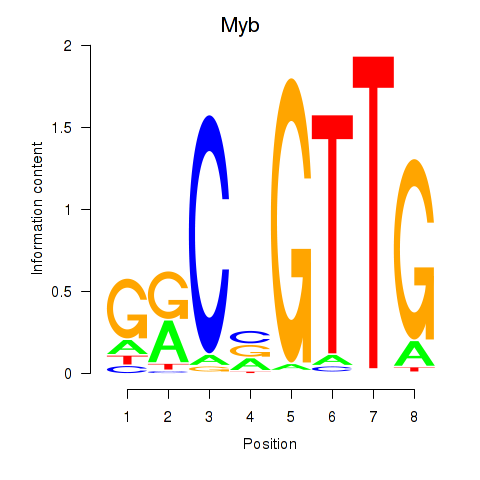
Activity profile of Myb motif
Sorted Z-values of Myb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Myb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_66315144 | 47.80 |
ENSMUST00000022618.6
|
Adam2
|
a disintegrin and metallopeptidase domain 2 |
| chr8_+_84116507 | 42.43 |
ENSMUST00000109831.3
|
Clgn
|
calmegin |
| chr14_+_70077841 | 41.81 |
ENSMUST00000022678.5
|
Pebp4
|
phosphatidylethanolamine binding protein 4 |
| chr8_+_84116463 | 39.14 |
ENSMUST00000002259.13
|
Clgn
|
calmegin |
| chr7_+_15795735 | 31.66 |
ENSMUST00000209369.2
|
Zfp541
|
zinc finger protein 541 |
| chr8_-_73302068 | 30.17 |
ENSMUST00000058534.7
|
Med26
|
mediator complex subunit 26 |
| chr4_-_118294521 | 26.99 |
ENSMUST00000006565.13
|
Cdc20
|
cell division cycle 20 |
| chr2_+_29780532 | 26.55 |
ENSMUST00000113764.4
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr6_+_145561483 | 26.31 |
ENSMUST00000087445.7
|
Tuba3b
|
tubulin, alpha 3B |
| chr3_+_116388600 | 25.91 |
ENSMUST00000198386.5
ENSMUST00000198311.5 ENSMUST00000197335.2 |
Sass6
|
SAS-6 centriolar assembly protein |
| chr4_-_49681954 | 25.53 |
ENSMUST00000029991.3
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| chr17_-_5468938 | 25.49 |
ENSMUST00000189788.2
|
Ldhal6b
|
lactate dehydrogenase A-like 6B |
| chr17_-_9888639 | 25.16 |
ENSMUST00000057190.4
|
Pabpc6
|
poly(A) binding protein, cytoplasmic 6 |
| chr15_+_4928624 | 24.77 |
ENSMUST00000045736.9
|
Mroh2b
|
maestro heat-like repeat family member 2B |
| chr15_-_74508197 | 24.76 |
ENSMUST00000023271.8
|
Mroh4
|
maestro heat-like repeat family member 4 |
| chr9_+_107424488 | 24.26 |
ENSMUST00000010188.9
|
Zmynd10
|
zinc finger, MYND domain containing 10 |
| chr14_-_30973164 | 23.68 |
ENSMUST00000226565.2
ENSMUST00000022459.5 |
Phf7
|
PHD finger protein 7 |
| chr16_+_17026467 | 23.65 |
ENSMUST00000169803.5
|
Rimbp3
|
RIMS binding protein 3 |
| chr11_+_69826603 | 22.96 |
ENSMUST00000018698.12
|
Ybx2
|
Y box protein 2 |
| chr2_+_118644717 | 22.54 |
ENSMUST00000028803.14
ENSMUST00000126045.8 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr1_-_166066298 | 22.31 |
ENSMUST00000038782.4
ENSMUST00000194057.6 |
Mael
|
maelstrom spermatogenic transposon silencer |
| chr8_-_87307294 | 22.26 |
ENSMUST00000131423.8
ENSMUST00000152438.2 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr2_-_127673738 | 22.07 |
ENSMUST00000028858.8
|
Bub1
|
BUB1, mitotic checkpoint serine/threonine kinase |
| chr1_+_88154727 | 21.75 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr8_-_25215778 | 21.68 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr18_+_52826752 | 21.58 |
ENSMUST00000066193.5
|
Gykl1
|
glycerol kinase-like 1 |
| chr11_+_69826719 | 21.39 |
ENSMUST00000149194.8
|
Ybx2
|
Y box protein 2 |
| chr8_-_25215856 | 21.18 |
ENSMUST00000033958.15
|
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr8_-_25164767 | 20.97 |
ENSMUST00000033957.12
|
Adam18
|
a disintegrin and metallopeptidase domain 18 |
| chr2_-_157121440 | 20.88 |
ENSMUST00000143663.2
|
Mroh8
|
maestro heat-like repeat family member 8 |
| chr9_+_72345801 | 20.62 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr11_+_87507977 | 20.62 |
ENSMUST00000093956.4
|
Hsf5
|
heat shock transcription factor family member 5 |
| chr1_+_153300874 | 20.58 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr4_-_107541421 | 20.32 |
ENSMUST00000069271.5
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr6_+_124806541 | 20.32 |
ENSMUST00000024270.14
|
Cdca3
|
cell division cycle associated 3 |
| chr2_+_118644675 | 20.24 |
ENSMUST00000110842.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr1_-_156131155 | 20.17 |
ENSMUST00000141760.4
ENSMUST00000121146.10 |
Tdrd5
|
tudor domain containing 5 |
| chr1_+_135060431 | 20.15 |
ENSMUST00000187985.7
ENSMUST00000049449.11 |
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr1_+_134890288 | 20.00 |
ENSMUST00000027687.8
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr1_-_156130918 | 19.90 |
ENSMUST00000167528.9
|
Tdrd5
|
tudor domain containing 5 |
| chr6_+_124806506 | 19.86 |
ENSMUST00000150120.8
|
Cdca3
|
cell division cycle associated 3 |
| chr6_+_116241146 | 19.81 |
ENSMUST00000112900.9
ENSMUST00000036503.14 ENSMUST00000223495.2 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chr9_+_108270020 | 19.66 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr17_+_80514889 | 19.59 |
ENSMUST00000134652.2
|
Ttc39d
|
tetratricopeptide repeat domain 39D |
| chr2_-_37593287 | 19.45 |
ENSMUST00000072186.12
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr3_+_40663285 | 19.13 |
ENSMUST00000091184.9
|
Slc25a31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr5_+_123887759 | 18.09 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr9_+_106158549 | 18.05 |
ENSMUST00000191434.2
|
Poc1a
|
POC1 centriolar protein A |
| chr5_-_135378896 | 17.93 |
ENSMUST00000201534.2
ENSMUST00000044972.11 |
Fkbp6
|
FK506 binding protein 6 |
| chr2_+_118644475 | 17.63 |
ENSMUST00000134661.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr13_+_22165687 | 17.61 |
ENSMUST00000017126.6
|
Pom121l2
|
POM121 membrane glycoprotein-like 2 (rat) |
| chr13_-_26954110 | 17.57 |
ENSMUST00000055915.6
|
Hdgfl1
|
HDGF like 1 |
| chr16_-_64672011 | 17.03 |
ENSMUST00000207826.2
|
Zfp654
|
zinc finger protein 654 |
| chr19_-_5452521 | 16.92 |
ENSMUST00000235569.2
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr2_+_122479770 | 16.71 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr9_+_108269992 | 16.41 |
ENSMUST00000192995.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr7_-_28913382 | 16.17 |
ENSMUST00000169143.8
ENSMUST00000047846.13 |
Catsperg1
|
cation channel sperm associated auxiliary subunit gamma 1 |
| chr11_+_85061922 | 16.15 |
ENSMUST00000018623.4
|
1700125H20Rik
|
RIKEN cDNA 1700125H20 gene |
| chr11_+_62737936 | 16.10 |
ENSMUST00000150989.8
ENSMUST00000176577.2 |
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr5_+_3707171 | 16.06 |
ENSMUST00000198739.5
|
Tmbim7
|
transmembrane BAX inhibitor motif containing 7 |
| chr11_-_94376158 | 15.82 |
ENSMUST00000041705.8
|
Spata20
|
spermatogenesis associated 20 |
| chr2_-_180284468 | 15.69 |
ENSMUST00000037877.11
|
Tcfl5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr11_-_69871320 | 15.64 |
ENSMUST00000143175.2
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr5_-_135378729 | 15.62 |
ENSMUST00000201784.4
ENSMUST00000201791.4 |
Fkbp6
|
FK506 binding protein 6 |
| chr7_+_43885573 | 15.58 |
ENSMUST00000223070.2
ENSMUST00000205530.2 |
Gm36864
|
predicted gene, 36864 |
| chr11_-_82655132 | 15.58 |
ENSMUST00000021040.10
ENSMUST00000100722.5 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chr11_-_33463722 | 15.57 |
ENSMUST00000102815.10
|
Ranbp17
|
RAN binding protein 17 |
| chr19_-_29302680 | 15.48 |
ENSMUST00000052380.5
|
Insl6
|
insulin-like 6 |
| chr15_+_89452529 | 15.30 |
ENSMUST00000023295.3
ENSMUST00000230538.2 ENSMUST00000230978.2 |
Acr
|
acrosin prepropeptide |
| chr16_-_50411484 | 15.30 |
ENSMUST00000062439.6
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr7_-_43885552 | 15.22 |
ENSMUST00000236952.2
|
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr9_+_106158212 | 14.96 |
ENSMUST00000072206.14
|
Poc1a
|
POC1 centriolar protein A |
| chr5_+_33815466 | 14.73 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr19_+_41970148 | 14.65 |
ENSMUST00000026170.3
|
Ubtd1
|
ubiquitin domain containing 1 |
| chr1_-_156766957 | 14.58 |
ENSMUST00000171292.8
ENSMUST00000063199.13 ENSMUST00000027886.14 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr14_+_46997984 | 14.55 |
ENSMUST00000067426.6
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr7_+_28140352 | 14.52 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr5_-_8472696 | 14.46 |
ENSMUST00000171808.8
|
Dbf4
|
DBF4 zinc finger |
| chr4_-_83404690 | 14.43 |
ENSMUST00000107214.9
ENSMUST00000107215.9 ENSMUST00000030207.15 |
Psip1
|
PC4 and SFRS1 interacting protein 1 |
| chr8_-_106140106 | 14.36 |
ENSMUST00000167294.8
ENSMUST00000063071.13 |
Kctd19
|
potassium channel tetramerisation domain containing 19 |
| chr2_-_172212426 | 14.26 |
ENSMUST00000109139.8
ENSMUST00000028997.8 ENSMUST00000109140.10 |
Aurka
|
aurora kinase A |
| chr10_+_100426346 | 13.96 |
ENSMUST00000218464.2
ENSMUST00000188930.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr11_-_118021460 | 13.92 |
ENSMUST00000132685.9
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr3_-_88204286 | 13.84 |
ENSMUST00000107556.10
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr4_-_44167987 | 13.78 |
ENSMUST00000143337.2
|
Rnf38
|
ring finger protein 38 |
| chr11_+_62737887 | 13.67 |
ENSMUST00000036085.11
|
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr6_-_47571901 | 13.65 |
ENSMUST00000081721.13
ENSMUST00000114618.8 ENSMUST00000114616.8 |
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr12_+_76450941 | 13.61 |
ENSMUST00000080449.7
|
Hspa2
|
heat shock protein 2 |
| chr9_+_30941924 | 13.46 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr16_+_16888145 | 13.46 |
ENSMUST00000232574.2
|
Ypel1
|
yippee like 1 |
| chr5_+_108280668 | 13.24 |
ENSMUST00000047677.9
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr19_-_9112919 | 13.22 |
ENSMUST00000049948.6
|
Asrgl1
|
asparaginase like 1 |
| chr11_+_102556397 | 13.17 |
ENSMUST00000100378.4
|
Meioc
|
meiosis specific with coiled-coil domain |
| chr17_-_34109513 | 13.16 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr2_-_125993887 | 13.15 |
ENSMUST00000110448.3
ENSMUST00000110446.9 |
Fam227b
|
family with sequence similarity 227, member B |
| chr1_+_6285082 | 13.12 |
ENSMUST00000160062.8
|
Rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr1_-_191307648 | 13.10 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase |
| chr12_-_79239022 | 13.08 |
ENSMUST00000161204.8
|
Rdh11
|
retinol dehydrogenase 11 |
| chr7_-_97387145 | 12.99 |
ENSMUST00000084986.8
|
Aqp11
|
aquaporin 11 |
| chr7_-_43885522 | 12.91 |
ENSMUST00000206686.2
ENSMUST00000037220.5 |
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr3_-_88204145 | 12.83 |
ENSMUST00000010682.4
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr3_+_103821413 | 12.82 |
ENSMUST00000051139.13
ENSMUST00000068879.11 |
Rsbn1
|
rosbin, round spermatid basic protein 1 |
| chr8_+_104977493 | 12.78 |
ENSMUST00000034342.13
ENSMUST00000212433.2 ENSMUST00000211809.2 |
Cklf
|
chemokine-like factor |
| chr1_-_131066004 | 12.72 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr3_-_51248032 | 12.59 |
ENSMUST00000062009.14
ENSMUST00000194641.6 |
Elf2
|
E74-like factor 2 |
| chr1_-_44141574 | 12.58 |
ENSMUST00000143327.2
ENSMUST00000133677.8 ENSMUST00000129702.2 ENSMUST00000149502.8 ENSMUST00000156392.8 ENSMUST00000150911.8 |
Tex30
|
testis expressed 30 |
| chr8_-_85696369 | 12.56 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr6_+_125026865 | 12.54 |
ENSMUST00000112413.8
|
Acrbp
|
proacrosin binding protein |
| chr6_-_131365380 | 12.42 |
ENSMUST00000032309.13
ENSMUST00000087865.4 |
Ybx3
|
Y box protein 3 |
| chr1_+_87111033 | 12.31 |
ENSMUST00000044533.9
|
Prss56
|
protease, serine 56 |
| chr4_+_110254907 | 12.30 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr9_-_108444561 | 12.29 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr1_+_135060994 | 12.29 |
ENSMUST00000167080.3
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr13_+_22165363 | 12.25 |
ENSMUST00000117882.2
|
Pom121l2
|
POM121 membrane glycoprotein-like 2 (rat) |
| chr11_+_45871135 | 12.18 |
ENSMUST00000049038.4
|
Sox30
|
SRY (sex determining region Y)-box 30 |
| chr5_-_8472582 | 12.15 |
ENSMUST00000168500.8
ENSMUST00000002368.16 |
Dbf4
|
DBF4 zinc finger |
| chr11_-_4390745 | 12.14 |
ENSMUST00000109948.8
|
Hormad2
|
HORMA domain containing 2 |
| chr1_-_156767123 | 12.02 |
ENSMUST00000189316.7
ENSMUST00000190648.7 ENSMUST00000172057.8 ENSMUST00000191605.7 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr11_+_104468107 | 11.95 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr1_-_44141503 | 11.91 |
ENSMUST00000128190.8
ENSMUST00000147571.8 ENSMUST00000027215.12 ENSMUST00000147661.8 |
Tex30
|
testis expressed 30 |
| chr17_+_56935118 | 11.90 |
ENSMUST00000112979.4
|
Catsperd
|
cation channel sperm associated auxiliary subunit delta |
| chr19_-_41790458 | 11.70 |
ENSMUST00000026150.15
ENSMUST00000163265.9 ENSMUST00000177495.2 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr12_+_76451177 | 11.59 |
ENSMUST00000219555.2
|
Hspa2
|
heat shock protein 2 |
| chr13_+_51799268 | 11.57 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr12_+_101370932 | 11.52 |
ENSMUST00000055156.5
|
Catsperb
|
cation channel sperm associated auxiliary subunit beta |
| chr8_-_106052884 | 11.51 |
ENSMUST00000210412.2
ENSMUST00000210801.2 ENSMUST00000070508.8 |
Lrrc29
|
leucine rich repeat containing 29 |
| chr18_+_34758062 | 11.50 |
ENSMUST00000166044.3
|
Kif20a
|
kinesin family member 20A |
| chr16_+_16888084 | 11.46 |
ENSMUST00000231514.2
|
Ypel1
|
yippee like 1 |
| chr10_+_127512933 | 11.41 |
ENSMUST00000118612.8
ENSMUST00000048099.5 |
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr6_-_125168637 | 11.13 |
ENSMUST00000043848.11
|
Ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr5_+_7354113 | 11.11 |
ENSMUST00000088796.3
|
Tex47
|
testis expressed 47 |
| chr7_-_126736979 | 11.09 |
ENSMUST00000049931.6
|
Spn
|
sialophorin |
| chr5_+_7354130 | 11.08 |
ENSMUST00000160634.2
ENSMUST00000159546.2 |
Tex47
|
testis expressed 47 |
| chr17_+_71859026 | 11.06 |
ENSMUST00000124001.8
ENSMUST00000167641.8 ENSMUST00000064420.12 |
Spdya
|
speedy/RINGO cell cycle regulator family, member A |
| chr13_+_34918820 | 11.03 |
ENSMUST00000039605.8
|
Fam50b
|
family with sequence similarity 50, member B |
| chr1_+_6284823 | 10.88 |
ENSMUST00000027040.13
|
Rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr7_+_118311740 | 10.84 |
ENSMUST00000106557.8
|
Ccp110
|
centriolar coiled coil protein 110 |
| chr3_+_137570334 | 10.82 |
ENSMUST00000174561.8
ENSMUST00000173790.8 |
H2az1
|
H2A.Z variant histone 1 |
| chr16_+_16887991 | 10.82 |
ENSMUST00000232258.2
|
Ypel1
|
yippee like 1 |
| chr11_+_104467791 | 10.80 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chr11_-_4391082 | 10.77 |
ENSMUST00000109949.8
ENSMUST00000130174.2 |
Hormad2
|
HORMA domain containing 2 |
| chr14_+_26414422 | 10.52 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr10_+_84674008 | 10.50 |
ENSMUST00000095388.5
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr12_+_103277234 | 10.46 |
ENSMUST00000191218.7
|
Fam181a
|
family with sequence similarity 181, member A |
| chr3_+_137570248 | 10.41 |
ENSMUST00000041045.14
|
H2az1
|
H2A.Z variant histone 1 |
| chr5_+_33815910 | 10.40 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr8_+_104977575 | 10.20 |
ENSMUST00000212939.2
|
Cklf
|
chemokine-like factor |
| chr1_-_156767196 | 10.13 |
ENSMUST00000185198.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr7_-_30298287 | 10.11 |
ENSMUST00000108150.2
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr14_+_63673843 | 10.08 |
ENSMUST00000121288.2
|
Fam167a
|
family with sequence similarity 167, member A |
| chr7_-_45084012 | 10.01 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr2_-_153286361 | 9.95 |
ENSMUST00000109784.2
|
Nol4l
|
nucleolar protein 4-like |
| chr4_+_148025316 | 9.88 |
ENSMUST00000103232.2
|
2510039O18Rik
|
RIKEN cDNA 2510039O18 gene |
| chr7_+_12758046 | 9.85 |
ENSMUST00000005705.8
|
Trim28
|
tripartite motif-containing 28 |
| chr13_-_43634695 | 9.61 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr7_-_97387429 | 9.59 |
ENSMUST00000206389.2
|
Aqp11
|
aquaporin 11 |
| chr10_-_41685603 | 9.56 |
ENSMUST00000190022.7
ENSMUST00000019951.16 ENSMUST00000186239.7 ENSMUST00000191498.2 |
Cep57l1
|
centrosomal protein 57-like 1 |
| chr7_-_44854316 | 9.52 |
ENSMUST00000121017.5
|
Kash5
|
KASH domain containing 5 |
| chr4_-_129636073 | 9.51 |
ENSMUST00000066257.6
|
Khdrbs1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr1_-_10079325 | 9.44 |
ENSMUST00000176398.8
ENSMUST00000027049.10 |
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr18_+_34757666 | 9.44 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr1_-_131065967 | 9.32 |
ENSMUST00000189756.2
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr3_+_30910089 | 9.26 |
ENSMUST00000108261.8
ENSMUST00000108259.8 ENSMUST00000166278.7 ENSMUST00000046748.13 ENSMUST00000194979.6 |
Gpr160
|
G protein-coupled receptor 160 |
| chr11_+_23615612 | 9.23 |
ENSMUST00000109525.8
ENSMUST00000020520.11 |
Pus10
|
pseudouridylate synthase 10 |
| chr1_+_58035130 | 9.22 |
ENSMUST00000027202.9
|
Sgo2a
|
shugoshin 2A |
| chr3_+_146110387 | 9.11 |
ENSMUST00000106151.8
ENSMUST00000106153.9 ENSMUST00000039021.11 ENSMUST00000106149.8 ENSMUST00000149262.8 |
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr14_+_46998004 | 9.07 |
ENSMUST00000227149.2
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr10_+_80100812 | 9.06 |
ENSMUST00000105362.8
ENSMUST00000105361.10 |
Dazap1
|
DAZ associated protein 1 |
| chr6_+_29402868 | 9.04 |
ENSMUST00000154619.5
|
Ccdc136
|
coiled-coil domain containing 136 |
| chr17_+_27136065 | 9.01 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr10_-_41685864 | 9.00 |
ENSMUST00000189770.7
ENSMUST00000187143.7 |
Cep57l1
|
centrosomal protein 57-like 1 |
| chr3_+_14598848 | 8.95 |
ENSMUST00000108370.9
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr3_+_14598877 | 8.89 |
ENSMUST00000169079.8
ENSMUST00000091325.10 |
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr9_-_35469818 | 8.85 |
ENSMUST00000034612.7
|
Ddx25
|
DEAD box helicase 25 |
| chr7_+_126461601 | 8.78 |
ENSMUST00000132808.2
|
Hirip3
|
HIRA interacting protein 3 |
| chr17_-_7228555 | 8.76 |
ENSMUST00000063683.8
|
Tagap1
|
T cell activation GTPase activating protein 1 |
| chr7_+_28140450 | 8.71 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr7_-_45083688 | 8.63 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr3_+_14598927 | 8.57 |
ENSMUST00000163660.8
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr3_+_30910163 | 8.50 |
ENSMUST00000108258.8
ENSMUST00000147697.2 |
Gpr160
|
G protein-coupled receptor 160 |
| chr4_-_133954669 | 8.49 |
ENSMUST00000105878.3
ENSMUST00000055892.10 ENSMUST00000169381.8 |
Catsper4
|
cation channel, sperm associated 4 |
| chr15_-_75941530 | 8.47 |
ENSMUST00000002603.12
ENSMUST00000063747.12 ENSMUST00000109946.9 |
Scrib
|
scribbled planar cell polarity |
| chr3_+_146110709 | 8.36 |
ENSMUST00000129978.2
|
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr9_-_97252011 | 8.27 |
ENSMUST00000035026.5
|
Trim42
|
tripartite motif-containing 42 |
| chr12_-_85327136 | 8.26 |
ENSMUST00000065913.8
ENSMUST00000008966.13 |
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr18_+_56840813 | 8.25 |
ENSMUST00000025486.9
|
Lmnb1
|
lamin B1 |
| chr4_+_110254858 | 8.23 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr14_-_119160497 | 8.23 |
ENSMUST00000047208.12
|
Dzip1
|
DAZ interacting protein 1 |
| chrX_+_133208833 | 8.20 |
ENSMUST00000081064.12
ENSMUST00000101251.8 ENSMUST00000129782.2 |
Cenpi
|
centromere protein I |
| chr9_+_107869662 | 8.08 |
ENSMUST00000177173.8
|
Cdhr4
|
cadherin-related family member 4 |
| chr9_-_121668527 | 8.03 |
ENSMUST00000135986.9
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr5_-_122510292 | 8.00 |
ENSMUST00000031419.6
|
Fam216a
|
family with sequence similarity 216, member A |
| chr11_+_82782938 | 7.96 |
ENSMUST00000018988.6
|
Fndc8
|
fibronectin type III domain containing 8 |
| chr6_+_83133381 | 7.86 |
ENSMUST00000032106.6
|
1700003E16Rik
|
RIKEN cDNA 1700003E16 gene |
| chr4_-_44167509 | 7.84 |
ENSMUST00000098098.9
|
Rnf38
|
ring finger protein 38 |
| chr12_-_84195222 | 7.83 |
ENSMUST00000061425.3
|
Pnma1
|
paraneoplastic antigen MA1 |
| chr15_-_36609208 | 7.80 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr18_+_34757687 | 7.77 |
ENSMUST00000237407.2
|
Kif20a
|
kinesin family member 20A |
| chr6_-_28261881 | 7.77 |
ENSMUST00000115320.8
ENSMUST00000123098.8 ENSMUST00000115321.9 ENSMUST00000155494.2 |
Zfp800
|
zinc finger protein 800 |
| chr14_-_62998561 | 7.67 |
ENSMUST00000053959.7
ENSMUST00000223585.2 |
Ints6
|
integrator complex subunit 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.4 | 40.1 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 8.4 | 25.2 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 8.0 | 24.0 | GO:0061723 | glycophagy(GO:0061723) |
| 7.7 | 23.2 | GO:0071846 | actin filament debranching(GO:0071846) |
| 6.3 | 31.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 5.6 | 22.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 5.4 | 27.0 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 5.3 | 26.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 4.8 | 14.3 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 4.5 | 13.6 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 4.4 | 22.0 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 4.4 | 13.2 | GO:0006530 | asparagine catabolic process(GO:0006530) |
| 4.3 | 21.6 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 4.1 | 33.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 4.1 | 8.2 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 4.1 | 20.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 4.0 | 20.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 3.8 | 11.5 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 3.7 | 11.1 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 3.7 | 36.7 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 3.4 | 60.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 3.3 | 13.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 3.3 | 13.1 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 3.2 | 9.5 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 3.1 | 18.6 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 3.1 | 12.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 3.0 | 27.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 2.9 | 22.9 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 2.7 | 18.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 2.5 | 140.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 2.5 | 55.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 2.5 | 7.6 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 2.5 | 7.6 | GO:0072708 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 2.5 | 9.8 | GO:1901536 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) positive regulation of methylation-dependent chromatin silencing(GO:0090309) negative regulation of DNA demethylation(GO:1901536) |
| 2.3 | 6.8 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 2.2 | 15.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 2.1 | 12.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 2.1 | 12.4 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) positive regulation of protein localization to centrosome(GO:1904781) |
| 2.0 | 34.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 1.9 | 21.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 1.9 | 5.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 1.8 | 18.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.8 | 10.8 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 1.8 | 10.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 1.7 | 5.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 1.6 | 13.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.6 | 25.5 | GO:0007320 | insemination(GO:0007320) |
| 1.6 | 10.9 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 1.5 | 4.6 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 1.5 | 7.4 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 1.5 | 4.4 | GO:0061511 | centriole elongation(GO:0061511) |
| 1.5 | 14.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 1.4 | 7.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 1.3 | 5.4 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 1.3 | 30.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 1.3 | 6.4 | GO:0071033 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 1.3 | 8.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 1.2 | 28.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 1.2 | 8.5 | GO:0016080 | synaptic vesicle targeting(GO:0016080) cochlear nucleus development(GO:0021747) |
| 1.2 | 29.6 | GO:0007099 | centriole replication(GO:0007099) |
| 1.2 | 5.8 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.1 | 11.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 1.1 | 18.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 1.1 | 6.4 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 1.1 | 30.6 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 1.0 | 11.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 1.0 | 196.2 | GO:0007286 | spermatid development(GO:0007286) |
| 1.0 | 3.8 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 1.0 | 12.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.9 | 2.8 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.8 | 8.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.8 | 13.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.8 | 2.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.8 | 13.3 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.8 | 7.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.8 | 3.0 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.7 | 8.7 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.7 | 5.8 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.7 | 8.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.7 | 2.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.7 | 6.2 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.7 | 2.7 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.7 | 2.6 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.6 | 4.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.6 | 4.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.6 | 20.1 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.6 | 7.9 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.6 | 6.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.5 | 2.2 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.5 | 3.8 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.5 | 2.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.5 | 1.5 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.5 | 5.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.5 | 29.2 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.5 | 4.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.5 | 19.8 | GO:0003341 | cilium movement(GO:0003341) |
| 0.5 | 3.3 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.5 | 4.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.5 | 1.8 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.4 | 51.1 | GO:0008542 | visual learning(GO:0008542) |
| 0.4 | 3.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.4 | 7.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.4 | 22.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.4 | 1.6 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.4 | 6.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.4 | 5.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 3.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.4 | 23.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.4 | 1.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 | 3.8 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.4 | 4.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.3 | 13.6 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.3 | 2.7 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.3 | 1.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 5.1 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.3 | 2.7 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.3 | 5.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.3 | 19.0 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.3 | 2.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.3 | 8.2 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.3 | 12.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.3 | 1.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 2.8 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.3 | 1.0 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.3 | 6.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 111.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.2 | 5.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 5.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 4.0 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.2 | 7.4 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.2 | 1.3 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 1.8 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 0.9 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.2 | 3.7 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 3.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.2 | 10.3 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.2 | 3.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 28.7 | GO:0050657 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) |
| 0.2 | 2.7 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.2 | 12.6 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.2 | 4.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 5.8 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.2 | 14.1 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.2 | 16.6 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.2 | 1.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 4.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 7.8 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.1 | 0.3 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 2.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.5 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 1.9 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 11.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 3.0 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.6 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 10.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 9.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.9 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 2.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.5 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 3.2 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 6.8 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 1.5 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 8.2 | GO:0042098 | T cell proliferation(GO:0042098) |
| 0.0 | 3.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 20.1 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 1.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 1.6 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 1.1 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 4.6 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 3.6 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.2 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 4.1 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 4.8 | GO:0042593 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 3.8 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 3.8 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 1.6 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 3.8 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 6.7 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 2.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 5.5 | 22.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 5.2 | 73.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 5.0 | 40.1 | GO:0071546 | pi-body(GO:0071546) |
| 3.8 | 15.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 3.7 | 22.3 | GO:0071547 | piP-body(GO:0071547) |
| 3.7 | 25.9 | GO:0098536 | deuterosome(GO:0098536) |
| 3.7 | 18.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 3.6 | 14.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 3.0 | 33.1 | GO:0002177 | manchette(GO:0002177) |
| 2.4 | 24.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 2.3 | 20.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 2.2 | 15.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 2.1 | 12.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 2.1 | 68.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 2.1 | 18.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 1.9 | 13.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.8 | 34.8 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 1.7 | 8.5 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 1.4 | 66.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 1.4 | 5.6 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 1.3 | 3.8 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 1.3 | 16.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 1.2 | 7.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 1.2 | 8.2 | GO:0005638 | lamin filament(GO:0005638) |
| 1.1 | 19.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 1.1 | 12.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 1.1 | 10.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 1.0 | 5.2 | GO:0031251 | PAN complex(GO:0031251) |
| 1.0 | 20.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.9 | 4.5 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.9 | 16.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.9 | 15.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.9 | 15.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.9 | 12.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.8 | 85.2 | GO:0005814 | centriole(GO:0005814) |
| 0.8 | 19.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 1.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.8 | 5.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.8 | 5.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.8 | 9.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.8 | 58.1 | GO:0005844 | polysome(GO:0005844) |
| 0.8 | 57.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.7 | 3.6 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.7 | 13.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.7 | 8.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.7 | 19.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.7 | 21.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.6 | 20.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.6 | 3.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.6 | 30.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.6 | 3.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.6 | 7.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.6 | 5.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.6 | 38.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.5 | 7.9 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.5 | 51.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.5 | 2.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.5 | 6.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.4 | 7.6 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.4 | 0.9 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.4 | 3.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.4 | 3.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 27.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.4 | 56.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.3 | 50.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 3.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 3.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 2.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 4.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 5.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 29.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 16.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 2.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 5.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 1.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.3 | 9.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 4.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 6.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 6.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 2.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 2.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 2.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 79.2 | GO:0005813 | centrosome(GO:0005813) |
| 0.2 | 9.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 14.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.2 | 22.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 5.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 76.4 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 15.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 7.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 10.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 3.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 8.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 8.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.7 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 1.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 17.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 3.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 3.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 4.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 22.2 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 20.0 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 5.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 12.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 23.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 7.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 125.4 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.4 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 4.5 | 27.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 4.4 | 13.2 | GO:0004067 | asparaginase activity(GO:0004067) |
| 4.3 | 21.6 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 3.3 | 9.8 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 3.2 | 12.8 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 2.8 | 25.5 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 2.4 | 97.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 2.3 | 18.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 2.2 | 6.5 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 2.1 | 8.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.9 | 11.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.9 | 13.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.8 | 12.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.8 | 23.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.7 | 8.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 1.7 | 6.8 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 1.7 | 18.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.7 | 51.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 1.7 | 23.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.6 | 33.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.5 | 4.6 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 1.5 | 4.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 1.4 | 35.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 1.4 | 5.5 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 1.4 | 13.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.3 | 22.6 | GO:0015250 | water channel activity(GO:0015250) |
| 1.2 | 3.7 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 1.2 | 4.8 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 1.0 | 25.2 | GO:0051861 | glycolipid binding(GO:0051861) |
| 1.0 | 56.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.9 | 24.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.9 | 3.8 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.8 | 14.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.7 | 30.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.7 | 13.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.7 | 5.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.7 | 2.7 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.7 | 20.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.6 | 14.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.6 | 6.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.6 | 1.9 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.6 | 7.9 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.6 | 3.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 2.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.5 | 43.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.5 | 65.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.5 | 1.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.5 | 14.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 21.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.5 | 5.4 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.5 | 3.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.5 | 3.3 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.5 | 7.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.5 | 22.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 4.9 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 10.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 7.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 1.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.4 | 8.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 23.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.4 | 19.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 11.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 6.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 1.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 18.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 10.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 11.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 8.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 11.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.3 | 32.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.3 | 6.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.3 | 18.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.3 | 38.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.3 | 5.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 5.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 9.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 12.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 3.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 6.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 0.6 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.2 | 2.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 17.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 26.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 9.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 3.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 17.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 11.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 5.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.6 | GO:0004103 | choline kinase activity(GO:0004103) cholinesterase activity(GO:0004104) |
| 0.1 | 19.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 12.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 1.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 5.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 24.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 19.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 3.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 15.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 59.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 43.1 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 2.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 7.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 6.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 2.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 18.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.6 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 13.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 3.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 6.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 3.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 5.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 7.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 158.0 | GO:0003677 | DNA binding(GO:0003677) |
| 0.1 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 18.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 2.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 2.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.7 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 4.1 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 1.2 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 9.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 120.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 1.5 | 32.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 1.2 | 47.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.7 | 11.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.6 | 53.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.6 | 28.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.4 | 18.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 19.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.4 | 9.8 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 11.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 3.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 12.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 4.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 12.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 5.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 7.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 7.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 5.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 5.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 3.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 16.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 8.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 3.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 9.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 14.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 1.3 | 67.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 1.3 | 35.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 1.3 | 41.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.1 | 21.7 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.7 | 29.1 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.6 | 18.6 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.6 | 19.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.6 | 56.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.4 | 12.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.4 | 5.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.4 | 7.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.3 | 11.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 32.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.3 | 4.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 13.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 11.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 4.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 4.8 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.2 | 10.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 7.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 11.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 12.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 1.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 4.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 11.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 5.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 4.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 3.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 7.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 2.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 7.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 4.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 4.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 2.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 3.5 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |