Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
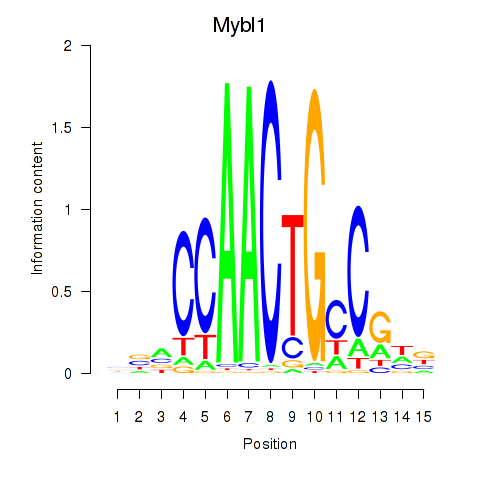
Results for Mybl1
Z-value: 1.81
Transcription factors associated with Mybl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mybl1
|
ENSMUSG00000025912.17 | Mybl1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl1 | mm39_v1_chr1_-_9770434_9770554 | 0.74 | 1.7e-13 | Click! |
Activity profile of Mybl1 motif
Sorted Z-values of Mybl1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Mybl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_30845909 | 14.39 |
ENSMUST00000236140.2
ENSMUST00000236118.2 ENSMUST00000235390.2 |
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr5_-_5529119 | 13.60 |
ENSMUST00000115447.2
|
Pttg1ip2
|
PTTG1IP family member 2 |
| chr11_+_98441923 | 12.03 |
ENSMUST00000081033.13
ENSMUST00000107511.8 ENSMUST00000107509.8 ENSMUST00000017339.12 |
Zpbp2
|
zona pellucida binding protein 2 |
| chr14_-_66315144 | 11.66 |
ENSMUST00000022618.6
|
Adam2
|
a disintegrin and metallopeptidase domain 2 |
| chr4_-_46413484 | 11.10 |
ENSMUST00000071096.3
|
Hemgn
|
hemogen |
| chr8_-_25215778 | 10.53 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr1_+_88154727 | 9.49 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr14_-_59416198 | 9.48 |
ENSMUST00000066558.4
|
Gm5142
|
predicted gene 5142 |
| chr1_-_156131155 | 9.33 |
ENSMUST00000141760.4
ENSMUST00000121146.10 |
Tdrd5
|
tudor domain containing 5 |
| chr15_-_74508197 | 9.27 |
ENSMUST00000023271.8
|
Mroh4
|
maestro heat-like repeat family member 4 |
| chr11_+_98441998 | 8.27 |
ENSMUST00000107513.3
|
Zpbp2
|
zona pellucida binding protein 2 |
| chr6_-_73446560 | 8.25 |
ENSMUST00000070163.6
|
4931417E11Rik
|
RIKEN cDNA 4931417E11 gene |
| chr7_-_43885522 | 8.13 |
ENSMUST00000206686.2
ENSMUST00000037220.5 |
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr7_+_106786300 | 8.02 |
ENSMUST00000142623.3
|
Nlrp14
|
NLR family, pyrin domain containing 14 |
| chr10_+_61007733 | 7.88 |
ENSMUST00000122261.8
ENSMUST00000121297.8 ENSMUST00000035894.12 |
Tbata
|
thymus, brain and testes associated |
| chr11_-_69709283 | 7.82 |
ENSMUST00000056941.3
|
Spem2
|
SPEM family member 2 |
| chr9_+_110673565 | 7.56 |
ENSMUST00000176403.8
|
Prss46
|
protease, serine 46 |
| chr8_-_25438784 | 7.55 |
ENSMUST00000119720.8
ENSMUST00000121438.9 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr1_+_186699613 | 7.51 |
ENSMUST00000045108.2
|
D1Pas1
|
DNA segment, Chr 1, Pasteur Institute 1 |
| chr7_-_43885552 | 7.43 |
ENSMUST00000236952.2
|
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr2_+_118644717 | 7.36 |
ENSMUST00000028803.14
ENSMUST00000126045.8 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr5_+_3707171 | 7.33 |
ENSMUST00000198739.5
|
Tmbim7
|
transmembrane BAX inhibitor motif containing 7 |
| chr13_+_34918820 | 7.32 |
ENSMUST00000039605.8
|
Fam50b
|
family with sequence similarity 50, member B |
| chr8_-_129791969 | 7.29 |
ENSMUST00000125112.8
ENSMUST00000108747.3 ENSMUST00000214889.2 ENSMUST00000095158.11 |
Ccdc7a
|
coiled-coil domain containing 7A |
| chr11_-_91468339 | 7.26 |
ENSMUST00000061019.6
|
Kif2b
|
kinesin family member 2B |
| chr8_-_106140106 | 7.19 |
ENSMUST00000167294.8
ENSMUST00000063071.13 |
Kctd19
|
potassium channel tetramerisation domain containing 19 |
| chr2_-_3420056 | 7.17 |
ENSMUST00000115084.8
ENSMUST00000115083.8 |
Meig1
|
meiosis expressed gene 1 |
| chr17_-_40519480 | 7.17 |
ENSMUST00000033585.7
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr2_-_3420102 | 7.14 |
ENSMUST00000115082.10
|
Meig1
|
meiosis expressed gene 1 |
| chr2_+_118644475 | 7.13 |
ENSMUST00000134661.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr11_+_58808830 | 7.13 |
ENSMUST00000020792.12
ENSMUST00000108818.4 |
Btnl10
|
butyrophilin-like 10 |
| chr2_+_150751475 | 7.08 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr10_+_40446326 | 7.00 |
ENSMUST00000078314.14
|
Slc22a16
|
solute carrier family 22 (organic cation transporter), member 16 |
| chr10_+_40446605 | 6.98 |
ENSMUST00000019978.9
|
Slc22a16
|
solute carrier family 22 (organic cation transporter), member 16 |
| chr9_+_108270020 | 6.93 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr16_+_9988080 | 6.91 |
ENSMUST00000121292.8
ENSMUST00000044103.6 |
Rpl39l
|
ribosomal protein L39-like |
| chr19_-_29302680 | 6.77 |
ENSMUST00000052380.5
|
Insl6
|
insulin-like 6 |
| chr7_+_43885573 | 6.73 |
ENSMUST00000223070.2
ENSMUST00000205530.2 |
Gm36864
|
predicted gene, 36864 |
| chrX_-_47966588 | 6.72 |
ENSMUST00000114928.2
|
Fsip2l
|
fibrous sheath-interacting protein 2-like |
| chr13_+_59860096 | 6.72 |
ENSMUST00000165133.3
|
Spata31d1b
|
spermatogenesis associated 31 subfamily D, member 1B |
| chr2_+_172821620 | 6.51 |
ENSMUST00000109125.8
ENSMUST00000109126.5 ENSMUST00000050442.15 |
Spo11
|
SPO11 initiator of meiotic double stranded breaks |
| chr5_-_143211632 | 6.48 |
ENSMUST00000085733.9
|
Spdye4a
|
speedy/RINGO cell cycle regulator family, member E4A |
| chr2_+_118644675 | 6.38 |
ENSMUST00000110842.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr19_-_11618165 | 6.20 |
ENSMUST00000186023.7
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr3_+_105821450 | 6.17 |
ENSMUST00000198080.5
ENSMUST00000199977.2 |
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr5_+_7354130 | 6.16 |
ENSMUST00000160634.2
ENSMUST00000159546.2 |
Tex47
|
testis expressed 47 |
| chr8_+_84682136 | 6.13 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr11_-_61233647 | 6.07 |
ENSMUST00000134423.3
ENSMUST00000093029.9 |
Slc47a2
|
solute carrier family 47, member 2 |
| chr1_-_59134042 | 6.03 |
ENSMUST00000238601.2
ENSMUST00000238949.2 ENSMUST00000097080.4 |
C2cd6
|
C2 calcium dependent domain containing 6 |
| chr1_+_153300874 | 5.94 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr15_-_88863210 | 5.93 |
ENSMUST00000042594.13
ENSMUST00000109368.2 |
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) |
| chr8_+_84116507 | 5.92 |
ENSMUST00000109831.3
|
Clgn
|
calmegin |
| chr8_+_96260694 | 5.91 |
ENSMUST00000041569.5
|
Ccdc113
|
coiled-coil domain containing 113 |
| chr19_-_11618192 | 5.89 |
ENSMUST00000112984.4
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr15_+_89452529 | 5.88 |
ENSMUST00000023295.3
ENSMUST00000230538.2 ENSMUST00000230978.2 |
Acr
|
acrosin prepropeptide |
| chr11_+_87684299 | 5.87 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr6_-_99703344 | 5.86 |
ENSMUST00000008273.8
ENSMUST00000101120.11 ENSMUST00000203738.2 |
Prok2
|
prokineticin 2 |
| chr3_-_124219673 | 5.86 |
ENSMUST00000029598.10
|
1700006A11Rik
|
RIKEN cDNA 1700006A11 gene |
| chr4_-_120966396 | 5.84 |
ENSMUST00000106268.4
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chr6_-_122778598 | 5.81 |
ENSMUST00000165884.8
|
Slc2a3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr8_+_46387647 | 5.79 |
ENSMUST00000095326.10
|
Ccdc110
|
coiled-coil domain containing 110 |
| chr9_-_109678685 | 5.78 |
ENSMUST00000112022.5
|
Camp
|
cathelicidin antimicrobial peptide |
| chr12_-_99849660 | 5.75 |
ENSMUST00000221929.2
ENSMUST00000046485.5 |
Efcab11
|
EF-hand calcium binding domain 11 |
| chr6_+_124806541 | 5.74 |
ENSMUST00000024270.14
|
Cdca3
|
cell division cycle associated 3 |
| chr14_+_46997984 | 5.68 |
ENSMUST00000067426.6
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr17_-_33358473 | 5.68 |
ENSMUST00000131954.2
ENSMUST00000053896.8 |
Morc2b
|
microrchidia 2B |
| chr17_-_79662514 | 5.67 |
ENSMUST00000068958.9
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr1_-_71142305 | 5.66 |
ENSMUST00000027393.8
|
Bard1
|
BRCA1 associated RING domain 1 |
| chr6_+_18866308 | 5.63 |
ENSMUST00000031489.10
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr3_-_68952057 | 5.63 |
ENSMUST00000107802.8
|
Trim59
|
tripartite motif-containing 59 |
| chr14_-_56339915 | 5.57 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr1_+_9978863 | 5.56 |
ENSMUST00000052843.12
ENSMUST00000171802.8 ENSMUST00000125294.9 ENSMUST00000140948.9 |
Mcmdc2
|
minichromosome maintenance domain containing 2 |
| chr6_+_124806506 | 5.54 |
ENSMUST00000150120.8
|
Cdca3
|
cell division cycle associated 3 |
| chr4_-_117039809 | 5.48 |
ENSMUST00000065896.9
|
Kif2c
|
kinesin family member 2C |
| chr2_+_122479770 | 5.47 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr19_+_7589890 | 5.43 |
ENSMUST00000025929.12
ENSMUST00000239394.2 ENSMUST00000148558.3 |
Plaat5
|
phospholipase A and acyltransferase 5 |
| chr6_+_18866339 | 5.40 |
ENSMUST00000115396.7
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr12_+_111937978 | 5.40 |
ENSMUST00000079009.11
|
Tdrd9
|
tudor domain containing 9 |
| chr1_-_36722195 | 5.40 |
ENSMUST00000170295.8
ENSMUST00000114981.3 |
Fam178b
|
family with sequence similarity 178, member B |
| chr8_+_84116463 | 5.38 |
ENSMUST00000002259.13
|
Clgn
|
calmegin |
| chr7_+_43600038 | 5.38 |
ENSMUST00000072204.5
|
Klk1b8
|
kallikrein 1-related peptidase b8 |
| chr7_-_30371021 | 5.36 |
ENSMUST00000051495.7
|
Pmis2
|
PMIS2 transmembrane protein |
| chr11_+_54194831 | 5.33 |
ENSMUST00000000145.12
ENSMUST00000138515.8 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr5_+_7354113 | 5.33 |
ENSMUST00000088796.3
|
Tex47
|
testis expressed 47 |
| chr10_-_39009910 | 5.27 |
ENSMUST00000135785.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr7_+_127475968 | 5.25 |
ENSMUST00000131000.2
|
Zfp646
|
zinc finger protein 646 |
| chr2_+_29014106 | 5.25 |
ENSMUST00000129544.8
|
Setx
|
senataxin |
| chr6_+_41118120 | 5.24 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chrX_+_9715942 | 5.23 |
ENSMUST00000057113.3
|
H2al3
|
H2A histone family member L3 |
| chr17_-_28299569 | 5.23 |
ENSMUST00000129046.9
ENSMUST00000043925.16 |
Tcp11
|
t-complex protein 11 |
| chr9_+_108269992 | 5.15 |
ENSMUST00000192995.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr11_-_72441054 | 5.15 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr19_-_44180881 | 5.12 |
ENSMUST00000042026.5
|
Pkd2l1
|
polycystic kidney disease 2-like 1 |
| chr6_+_146751598 | 5.07 |
ENSMUST00000032433.9
|
Smco2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr14_+_66043281 | 5.05 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr10_+_14581325 | 5.05 |
ENSMUST00000191238.7
ENSMUST00000190114.2 |
Nmbr
|
neuromedin B receptor |
| chr1_+_58035130 | 5.05 |
ENSMUST00000027202.9
|
Sgo2a
|
shugoshin 2A |
| chr8_+_70285133 | 5.04 |
ENSMUST00000081503.13
|
Pbx4
|
pre B cell leukemia homeobox 4 |
| chr6_-_139987135 | 5.03 |
ENSMUST00000032356.13
|
Plcz1
|
phospholipase C, zeta 1 |
| chr6_-_99703295 | 5.02 |
ENSMUST00000032152.14
|
Prok2
|
prokineticin 2 |
| chr11_-_118021460 | 4.96 |
ENSMUST00000132685.9
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr1_-_156130918 | 4.95 |
ENSMUST00000167528.9
|
Tdrd5
|
tudor domain containing 5 |
| chr5_+_128813232 | 4.94 |
ENSMUST00000086056.8
|
Piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr11_+_82782938 | 4.92 |
ENSMUST00000018988.6
|
Fndc8
|
fibronectin type III domain containing 8 |
| chr11_-_99045894 | 4.91 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr12_+_76450941 | 4.88 |
ENSMUST00000080449.7
|
Hspa2
|
heat shock protein 2 |
| chr9_+_72345801 | 4.86 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr4_-_56741398 | 4.84 |
ENSMUST00000095080.5
|
Actl7b
|
actin-like 7b |
| chr6_-_18109037 | 4.84 |
ENSMUST00000010940.11
|
Asz1
|
ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| chr4_-_63518986 | 4.83 |
ENSMUST00000210529.2
|
Gm11213
|
predicted gene 11213 |
| chr6_+_139987275 | 4.81 |
ENSMUST00000043797.6
|
Capza3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr11_+_45871135 | 4.80 |
ENSMUST00000049038.4
|
Sox30
|
SRY (sex determining region Y)-box 30 |
| chr8_-_85696369 | 4.77 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr13_-_26954110 | 4.76 |
ENSMUST00000055915.6
|
Hdgfl1
|
HDGF like 1 |
| chr19_+_4204605 | 4.76 |
ENSMUST00000061086.9
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr9_+_95739650 | 4.75 |
ENSMUST00000034980.9
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr3_+_30910089 | 4.67 |
ENSMUST00000108261.8
ENSMUST00000108259.8 ENSMUST00000166278.7 ENSMUST00000046748.13 ENSMUST00000194979.6 |
Gpr160
|
G protein-coupled receptor 160 |
| chr9_+_21437440 | 4.66 |
ENSMUST00000086361.12
ENSMUST00000173769.3 |
AB124611
|
cDNA sequence AB124611 |
| chr19_-_46661321 | 4.60 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr11_-_100595019 | 4.59 |
ENSMUST00000017974.13
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr3_-_68952030 | 4.59 |
ENSMUST00000136512.3
|
Trim59
|
tripartite motif-containing 59 |
| chr11_-_105347500 | 4.56 |
ENSMUST00000049995.10
ENSMUST00000100332.4 |
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr19_-_47825790 | 4.53 |
ENSMUST00000160247.3
|
Cfap43
|
cilia and flagella associated protein 43 |
| chr9_+_56344700 | 4.51 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr12_+_76451177 | 4.47 |
ENSMUST00000219555.2
|
Hspa2
|
heat shock protein 2 |
| chr19_+_4889394 | 4.42 |
ENSMUST00000088653.3
|
Ccdc87
|
coiled-coil domain containing 87 |
| chr4_-_129452180 | 4.39 |
ENSMUST00000067240.11
|
Lck
|
lymphocyte protein tyrosine kinase |
| chrX_+_49930311 | 4.38 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr4_-_129452148 | 4.33 |
ENSMUST00000167288.8
ENSMUST00000134336.3 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr12_-_87283780 | 4.33 |
ENSMUST00000221768.2
|
Noxred1
|
NADP+ dependent oxidoreductase domain containing 1 |
| chr7_-_126736979 | 4.31 |
ENSMUST00000049931.6
|
Spn
|
sialophorin |
| chr1_-_156766957 | 4.29 |
ENSMUST00000171292.8
ENSMUST00000063199.13 ENSMUST00000027886.14 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr1_-_156936197 | 4.28 |
ENSMUST00000187546.7
ENSMUST00000118207.8 ENSMUST00000027884.13 ENSMUST00000121911.8 |
Tex35
|
testis expressed 35 |
| chr11_+_3282424 | 4.28 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr7_+_12246415 | 4.26 |
ENSMUST00000032541.5
|
2900092C05Rik
|
RIKEN cDNA 2900092C05 gene |
| chr9_+_95739747 | 4.24 |
ENSMUST00000215311.2
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr8_+_27937128 | 4.21 |
ENSMUST00000209669.2
|
Gm45861
|
predicted gene 45861 |
| chr11_-_59678462 | 4.20 |
ENSMUST00000125307.2
|
Pld6
|
phospholipase D family, member 6 |
| chr13_-_23946359 | 4.16 |
ENSMUST00000091701.3
|
H3c1
|
H3 clustered histone 1 |
| chr17_+_37504783 | 4.13 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr8_-_25164767 | 4.10 |
ENSMUST00000033957.12
|
Adam18
|
a disintegrin and metallopeptidase domain 18 |
| chr4_-_43499608 | 4.04 |
ENSMUST00000136005.3
ENSMUST00000054538.13 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr1_-_16163506 | 4.04 |
ENSMUST00000145070.8
ENSMUST00000151004.2 |
4930444P10Rik
|
RIKEN cDNA 4930444P10 gene |
| chr17_+_56935118 | 4.02 |
ENSMUST00000112979.4
|
Catsperd
|
cation channel sperm associated auxiliary subunit delta |
| chr14_+_40794817 | 3.99 |
ENSMUST00000189865.7
|
Dydc1
|
DPY30 domain containing 1 |
| chr1_+_98348817 | 3.97 |
ENSMUST00000027575.13
ENSMUST00000160796.2 |
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr14_+_66043515 | 3.97 |
ENSMUST00000139644.2
|
Pbk
|
PDZ binding kinase |
| chr3_+_30910163 | 3.95 |
ENSMUST00000108258.8
ENSMUST00000147697.2 |
Gpr160
|
G protein-coupled receptor 160 |
| chr10_+_127512933 | 3.93 |
ENSMUST00000118612.8
ENSMUST00000048099.5 |
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr2_+_174292471 | 3.91 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr19_-_46661501 | 3.90 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr3_+_54600509 | 3.90 |
ENSMUST00000170552.6
|
Supt20
|
SPT20 SAGA complex component |
| chr11_-_59266483 | 3.89 |
ENSMUST00000061481.7
|
Prss38
|
protease, serine 38 |
| chr9_-_102503180 | 3.86 |
ENSMUST00000216281.2
ENSMUST00000093791.10 |
Cep63
|
centrosomal protein 63 |
| chr11_-_32150222 | 3.82 |
ENSMUST00000145401.8
ENSMUST00000142396.2 ENSMUST00000128311.8 |
Il9r
|
interleukin 9 receptor |
| chr2_+_125089110 | 3.81 |
ENSMUST00000082122.14
|
Dut
|
deoxyuridine triphosphatase |
| chr17_-_35394971 | 3.79 |
ENSMUST00000173324.8
|
Aif1
|
allograft inflammatory factor 1 |
| chr8_+_106434901 | 3.79 |
ENSMUST00000013302.7
ENSMUST00000211852.2 |
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr5_+_114991722 | 3.79 |
ENSMUST00000031547.12
|
4930519G04Rik
|
RIKEN cDNA 4930519G04 gene |
| chr5_+_123887759 | 3.78 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr2_+_109111083 | 3.73 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr14_+_22069877 | 3.73 |
ENSMUST00000161249.8
ENSMUST00000159777.8 ENSMUST00000162540.2 |
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr7_-_73663422 | 3.72 |
ENSMUST00000026896.10
ENSMUST00000191970.2 |
St8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr1_-_156767123 | 3.70 |
ENSMUST00000189316.7
ENSMUST00000190648.7 ENSMUST00000172057.8 ENSMUST00000191605.7 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr7_-_5128936 | 3.68 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr4_-_44167987 | 3.67 |
ENSMUST00000143337.2
|
Rnf38
|
ring finger protein 38 |
| chr1_-_60137294 | 3.64 |
ENSMUST00000141417.3
ENSMUST00000122038.8 |
Wdr12
|
WD repeat domain 12 |
| chr14_+_46998004 | 3.64 |
ENSMUST00000227149.2
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr15_-_78456898 | 3.62 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr14_+_71011744 | 3.61 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr11_+_68322945 | 3.58 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr10_-_22607136 | 3.57 |
ENSMUST00000238910.2
ENSMUST00000127698.8 |
ENSMUSG00000118528.2
Tbpl1
|
novel protein TATA box binding protein-like 1 |
| chr15_-_89310060 | 3.57 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr1_-_172085977 | 3.55 |
ENSMUST00000111243.2
|
Atp1a4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr4_+_111577172 | 3.54 |
ENSMUST00000038868.14
ENSMUST00000070513.13 ENSMUST00000153746.8 |
Spata6
|
spermatogenesis associated 6 |
| chr3_+_54600196 | 3.51 |
ENSMUST00000197502.5
ENSMUST00000200441.5 ENSMUST00000029315.13 ENSMUST00000199655.5 |
Supt20
|
SPT20 SAGA complex component |
| chr1_-_44141574 | 3.51 |
ENSMUST00000143327.2
ENSMUST00000133677.8 ENSMUST00000129702.2 ENSMUST00000149502.8 ENSMUST00000156392.8 ENSMUST00000150911.8 |
Tex30
|
testis expressed 30 |
| chr7_-_19133783 | 3.50 |
ENSMUST00000047170.10
ENSMUST00000108459.9 |
Klc3
|
kinesin light chain 3 |
| chr5_+_38377814 | 3.49 |
ENSMUST00000087514.9
ENSMUST00000130721.8 ENSMUST00000123207.8 ENSMUST00000132190.8 ENSMUST00000202506.2 ENSMUST00000152066.8 ENSMUST00000155300.8 |
Lyar
|
Ly1 antibody reactive clone |
| chr6_+_34757346 | 3.48 |
ENSMUST00000115016.8
ENSMUST00000115017.8 |
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr17_+_48554786 | 3.47 |
ENSMUST00000048065.6
|
Trem3
|
triggering receptor expressed on myeloid cells 3 |
| chr13_+_22165687 | 3.47 |
ENSMUST00000017126.6
|
Pom121l2
|
POM121 membrane glycoprotein-like 2 (rat) |
| chr5_+_66417488 | 3.46 |
ENSMUST00000031109.7
|
Nsun7
|
NOL1/NOP2/Sun domain family, member 7 |
| chr1_-_44141503 | 3.45 |
ENSMUST00000128190.8
ENSMUST00000147571.8 ENSMUST00000027215.12 ENSMUST00000147661.8 |
Tex30
|
testis expressed 30 |
| chr7_-_34974953 | 3.45 |
ENSMUST00000238407.2
|
Wdr88
|
WD repeat domain 88 |
| chr5_+_38377972 | 3.44 |
ENSMUST00000114106.8
|
Lyar
|
Ly1 antibody reactive clone |
| chr17_-_35265702 | 3.43 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr5_-_72910106 | 3.42 |
ENSMUST00000197313.5
ENSMUST00000198464.3 ENSMUST00000113604.10 |
Txk
|
TXK tyrosine kinase |
| chr4_-_44168252 | 3.42 |
ENSMUST00000145760.8
ENSMUST00000128426.8 |
Rnf38
|
ring finger protein 38 |
| chrX_+_99472511 | 3.42 |
ENSMUST00000060241.3
|
Otud6a
|
OTU domain containing 6A |
| chr11_+_121036969 | 3.41 |
ENSMUST00000039088.9
ENSMUST00000155694.2 |
Tex19.1
|
testis expressed gene 19.1 |
| chr16_+_45430743 | 3.41 |
ENSMUST00000161347.9
ENSMUST00000023339.5 |
Gcsam
|
germinal center associated, signaling and motility |
| chr11_-_34674677 | 3.40 |
ENSMUST00000093193.12
ENSMUST00000101365.9 |
Dock2
|
dedicator of cyto-kinesis 2 |
| chr6_-_128868068 | 3.40 |
ENSMUST00000178918.2
ENSMUST00000160290.8 |
BC035044
|
cDNA sequence BC035044 |
| chr10_+_100426346 | 3.39 |
ENSMUST00000218464.2
ENSMUST00000188930.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr11_+_54194624 | 3.37 |
ENSMUST00000093106.12
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr13_+_55612050 | 3.37 |
ENSMUST00000046533.9
|
Prr7
|
proline rich 7 (synaptic) |
| chr14_-_31299275 | 3.34 |
ENSMUST00000112027.9
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr11_+_32246489 | 3.34 |
ENSMUST00000093207.4
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr6_-_113577606 | 3.34 |
ENSMUST00000035870.5
|
Fancd2os
|
Fancd2 opposite strand |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.5 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 4.0 | 12.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 3.1 | 9.4 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 2.2 | 9.0 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 2.2 | 10.9 | GO:0060983 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) epicardium-derived cardiac vascular smooth muscle cell differentiation(GO:0060983) |
| 2.1 | 10.5 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 2.0 | 6.0 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.8 | 5.3 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 1.6 | 4.9 | GO:2000547 | regulation of tolerance induction to self antigen(GO:0002649) lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 1.4 | 4.3 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 1.3 | 2.7 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 1.3 | 7.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 1.3 | 5.2 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 1.3 | 10.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 1.3 | 5.1 | GO:0036446 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 1.3 | 14.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 1.3 | 3.8 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 1.2 | 4.9 | GO:0000239 | pachytene(GO:0000239) |
| 1.2 | 5.9 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 1.2 | 5.8 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 1.1 | 3.4 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 1.1 | 3.4 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 1.1 | 3.4 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 1.1 | 14.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 1.1 | 23.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 1.1 | 10.7 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 1.1 | 7.5 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) |
| 1.1 | 5.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 1.1 | 3.2 | GO:0010138 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 1.0 | 3.1 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 1.0 | 20.9 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.0 | 5.9 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 1.0 | 3.8 | GO:0006226 | dUMP biosynthetic process(GO:0006226) pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.9 | 4.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.9 | 5.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 4.6 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.9 | 9.0 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.9 | 2.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.9 | 2.6 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.9 | 5.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.9 | 3.4 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.8 | 2.5 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.8 | 2.5 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.8 | 3.2 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.8 | 4.6 | GO:1900245 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.8 | 2.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.7 | 2.2 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.7 | 5.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.7 | 5.8 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.7 | 0.7 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.7 | 2.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.7 | 8.7 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.7 | 3.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.6 | 7.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.6 | 5.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.6 | 1.9 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.6 | 6.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.6 | 6.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.6 | 1.8 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.6 | 28.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.6 | 1.8 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.6 | 1.8 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.6 | 6.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.6 | 5.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.6 | 2.9 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.6 | 2.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 7.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.5 | 3.3 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.5 | 10.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.5 | 3.7 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.5 | 1.6 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.5 | 2.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.5 | 3.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.5 | 5.7 | GO:0085020 | negative regulation of protein export from nucleus(GO:0046826) protein K6-linked ubiquitination(GO:0085020) |
| 0.5 | 4.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.5 | 6.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.5 | 3.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.5 | 1.5 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.5 | 3.9 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.5 | 4.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 5.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.5 | 8.5 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.5 | 1.4 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.5 | 1.4 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.5 | 1.4 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.4 | 5.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.4 | 3.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.4 | 2.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 2.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.4 | 3.6 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.4 | 1.6 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 | 1.2 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.4 | 2.4 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) positive regulation of protein localization to centrosome(GO:1904781) |
| 0.4 | 2.7 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.4 | 1.6 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 1.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.4 | 1.9 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.4 | 1.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.4 | 9.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.4 | 2.6 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.4 | 3.2 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.4 | 2.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 2.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 5.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 2.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.3 | 1.3 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.3 | 2.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 2.3 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.3 | 7.0 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.3 | 4.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 1.6 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.3 | 0.9 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.3 | 4.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.3 | 6.3 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.3 | 1.3 | GO:0042710 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.3 | 1.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.3 | 3.0 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.3 | 1.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.3 | 1.2 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.3 | 3.8 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) embryonic brain development(GO:1990403) |
| 0.3 | 2.3 | GO:0060022 | hard palate development(GO:0060022) |
| 0.3 | 2.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 3.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 2.3 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 1.1 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.3 | 6.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.3 | 1.9 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.3 | 4.9 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.3 | 1.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.3 | 0.5 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.3 | 0.8 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.3 | 1.6 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.3 | 0.8 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.3 | 1.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 2.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 1.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 1.5 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 | 6.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 | 3.6 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.2 | 0.5 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.2 | 0.9 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 6.8 | GO:0003341 | cilium movement(GO:0003341) |
| 0.2 | 1.5 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 2.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.2 | 1.5 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.2 | 3.8 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 2.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 4.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 2.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 10.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.2 | 1.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 1.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 17.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.2 | 0.8 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.2 | 1.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 2.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 3.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 3.0 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.2 | 1.9 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 0.8 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.2 | 2.0 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.2 | 2.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 0.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.2 | 1.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.5 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 2.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 0.9 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 2.6 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 12.8 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.1 | 1.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.9 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 5.7 | GO:0036474 | cell death in response to hydrogen peroxide(GO:0036474) regulation of hydrogen peroxide-induced cell death(GO:1903205) |
| 0.1 | 0.9 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.3 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.5 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 1.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 8.0 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 2.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 1.6 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.1 | 0.9 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 70.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 0.7 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.1 | 2.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 2.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.7 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 | 1.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 2.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.9 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.7 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 2.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 2.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 11.5 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.1 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 0.7 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.1 | 2.0 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 6.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.8 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 | 4.7 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 3.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 8.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 3.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 2.5 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.1 | 3.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 1.5 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 0.4 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 0.9 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.7 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 5.9 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 0.2 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.7 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 4.6 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.1 | 1.5 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 5.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 2.3 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 4.4 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.5 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 1.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.4 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.1 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 2.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 1.0 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 4.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.9 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.2 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.8 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.8 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 3.1 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.1 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.6 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 2.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 2.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.6 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 0.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.5 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 3.2 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.7 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.0 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 1.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.9 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 2.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 5.5 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 4.8 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 1.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 1.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 1.6 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 1.0 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) negative regulation of gene silencing(GO:0060969) |
| 0.0 | 1.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.3 | GO:0032367 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.1 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0000811 | GINS complex(GO:0000811) |
| 2.2 | 6.6 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 1.9 | 19.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 1.8 | 5.4 | GO:0071547 | piP-body(GO:0071547) |
| 1.7 | 19.1 | GO:0071546 | pi-body(GO:0071546) |
| 1.5 | 5.9 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.4 | 8.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.3 | 6.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.3 | 15.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.3 | 3.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.9 | 18.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.9 | 12.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.8 | 3.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.7 | 6.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.7 | 3.6 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.7 | 2.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.6 | 30.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.6 | 3.7 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.6 | 10.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.6 | 2.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.6 | 3.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.5 | 4.9 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 5.7 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.5 | 4.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 6.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.5 | 18.1 | GO:0001741 | XY body(GO:0001741) |
| 0.5 | 12.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.4 | 5.0 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.4 | 2.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.4 | 6.0 | GO:0000243 | commitment complex(GO:0000243) |
| 0.4 | 3.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.4 | 2.3 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.4 | 6.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.3 | 4.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.3 | 9.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 2.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 2.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 1.6 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.3 | 4.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 6.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.3 | 4.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 2.0 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.3 | 3.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.3 | 3.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 1.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.3 | 2.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 1.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 1.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 1.8 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 3.6 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 29.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.2 | 2.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 7.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 1.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 0.6 | GO:0005712 | chiasma(GO:0005712) |
| 0.2 | 2.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 3.0 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 1.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 12.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 5.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 0.9 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 1.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 12.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 3.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 5.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 2.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 17.1 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 1.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 2.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.4 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 0.9 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 2.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 2.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 2.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 3.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 1.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 4.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 3.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 5.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.0 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.5 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 4.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.8 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 4.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 7.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 4.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 5.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 6.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 5.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 4.1 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 1.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 4.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.6 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 5.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 2.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 2.9 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 10.5 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 3.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 7.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.2 | GO:0097526 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 19.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 4.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 6.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.0 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 3.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.9 | GO:0005694 | chromosome(GO:0005694) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 14.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 1.8 | 5.3 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 1.7 | 8.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.7 | 5.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 1.6 | 6.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 1.6 | 6.5 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.4 | 7.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 1.3 | 5.3 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 1.2 | 5.8 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 1.1 | 7.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.1 | 9.0 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 1.0 | 4.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 1.0 | 5.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 1.0 | 6.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.0 | 2.9 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.9 | 7.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.9 | 2.7 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.9 | 2.7 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.9 | 5.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.9 | 3.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.9 | 26.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.8 | 2.5 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.8 | 3.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.8 | 3.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.8 | 3.0 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.8 | 2.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.8 | 2.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.6 | 2.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.6 | 3.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.6 | 1.8 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.6 | 2.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.6 | 1.8 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.6 | 2.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.5 | 10.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.5 | 1.5 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.5 | 3.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.5 | 1.8 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.5 | 5.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.5 | 3.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.5 | 10.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 7.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.4 | 5.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.4 | 1.8 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.4 | 2.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 5.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.4 | 8.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.4 | 3.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.4 | 2.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.4 | 1.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.4 | 9.4 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.4 | 1.1 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.4 | 8.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 2.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.4 | 9.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.4 | 1.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 3.9 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.4 | 3.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.4 | 1.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.4 | 1.4 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.3 | 1.7 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.3 | 4.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.3 | 1.9 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 1.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 0.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 4.0 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.3 | 1.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 0.9 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.3 | 2.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 3.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 4.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 7.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.3 | 4.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 10.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 5.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 5.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 2.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 2.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 2.5 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 2.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 6.9 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.2 | 2.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 0.9 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 2.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 1.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 0.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 3.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 1.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 0.9 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 13.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 1.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 4.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 3.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 4.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.8 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 1.5 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 1.2 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 3.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.4 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.2 | 9.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 3.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.9 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 2.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.8 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.5 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.1 | 10.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.0 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.7 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 2.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.8 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 1.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 13.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 5.7 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 1.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 2.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 3.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.3 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 1.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 4.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 2.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 5.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 2.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 2.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 1.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 12.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 2.0 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 3.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.2 | GO:0004314 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.1 | 15.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 2.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 3.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.7 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 5.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 2.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 2.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 1.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 3.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 2.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 16.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.0 | 1.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0047238 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 1.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 2.0 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 6.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 2.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 4.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.7 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.5 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.0 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 5.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 2.6 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 2.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 5.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 1.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 4.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 2.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.5 | 20.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.3 | 1.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 11.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 4.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 8.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 4.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 3.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 5.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 8.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 6.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 6.8 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 3.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 4.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 14.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.8 | 8.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 9.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.6 | 20.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 8.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 7.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 16.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.4 | 2.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.4 | 6.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 5.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 10.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 4.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.3 | 6.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 2.7 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.3 | 3.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 5.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 8.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 1.9 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.2 | 5.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 7.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.2 | 9.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 16.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 6.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 1.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 4.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 6.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 1.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 7.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.9 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 2.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 5.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 7.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 2.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 4.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 7.3 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.1 | 2.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.5 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 2.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 2.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 1.6 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 2.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 4.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |