Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
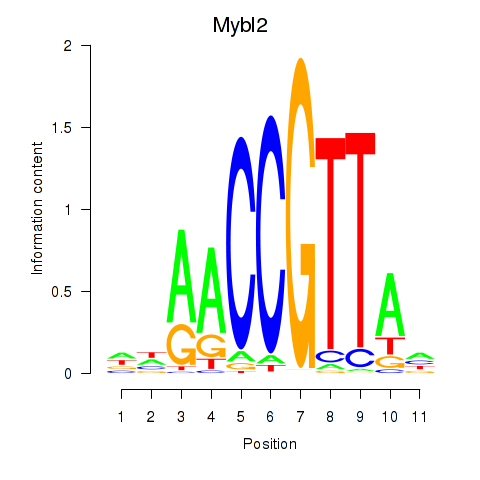
Results for Mybl2
Z-value: 1.89
Transcription factors associated with Mybl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mybl2
|
ENSMUSG00000017861.12 | Mybl2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl2 | mm39_v1_chr2_+_162916551_162916665 | 0.73 | 5.4e-13 | Click! |
Activity profile of Mybl2 motif
Sorted Z-values of Mybl2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Mybl2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_169358912 | 20.63 |
ENSMUST00000192248.2
ENSMUST00000028000.13 |
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr10_-_84938350 | 17.78 |
ENSMUST00000059383.8
ENSMUST00000216889.2 |
Fhl4
|
four and a half LIM domains 4 |
| chr2_+_29014106 | 13.70 |
ENSMUST00000129544.8
|
Setx
|
senataxin |
| chr1_-_169359015 | 13.52 |
ENSMUST00000111368.8
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr8_-_106140106 | 12.58 |
ENSMUST00000167294.8
ENSMUST00000063071.13 |
Kctd19
|
potassium channel tetramerisation domain containing 19 |
| chr15_+_26309125 | 12.34 |
ENSMUST00000126304.2
ENSMUST00000140840.8 ENSMUST00000152841.2 |
Marchf11
|
membrane associated ring-CH-type finger 11 |
| chr7_+_79944198 | 12.24 |
ENSMUST00000163812.9
ENSMUST00000047558.14 ENSMUST00000174199.8 ENSMUST00000173824.8 ENSMUST00000174172.8 |
Prc1
|
protein regulator of cytokinesis 1 |
| chr7_-_29979758 | 12.20 |
ENSMUST00000108190.8
|
Wdr62
|
WD repeat domain 62 |
| chr14_-_67953035 | 12.08 |
ENSMUST00000163100.8
ENSMUST00000132705.8 ENSMUST00000124045.3 |
Cdca2
|
cell division cycle associated 2 |
| chr6_+_41025217 | 11.41 |
ENSMUST00000103264.3
|
Trbv3
|
T cell receptor beta, variable 3 |
| chr1_+_191553556 | 11.02 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chrX_+_149330371 | 10.38 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr13_+_51799268 | 9.95 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr9_-_44891626 | 9.94 |
ENSMUST00000002101.12
ENSMUST00000160886.2 |
Cd3g
|
CD3 antigen, gamma polypeptide |
| chr3_-_102843406 | 9.73 |
ENSMUST00000199930.2
ENSMUST00000029448.11 ENSMUST00000196988.5 |
Sycp1
|
synaptonemal complex protein 1 |
| chr18_-_50701793 | 9.73 |
ENSMUST00000056460.4
|
Pudp
|
pseudouridine 5'-phosphatase |
| chr4_-_124830644 | 9.62 |
ENSMUST00000030690.12
ENSMUST00000084296.10 |
Cdca8
|
cell division cycle associated 8 |
| chr19_-_41790458 | 9.62 |
ENSMUST00000026150.15
ENSMUST00000163265.9 ENSMUST00000177495.2 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr3_+_108646974 | 9.05 |
ENSMUST00000133931.9
|
Aknad1
|
AKNA domain containing 1 |
| chr10_+_82702449 | 8.62 |
ENSMUST00000211623.2
|
Eid3
|
EP300 interacting inhibitor of differentiation 3 |
| chr16_-_18880821 | 8.62 |
ENSMUST00000200568.2
|
Iglc1
|
immunoglobulin lambda constant 1 |
| chr11_-_91468339 | 8.56 |
ENSMUST00000061019.6
|
Kif2b
|
kinesin family member 2B |
| chr9_+_108270020 | 8.51 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr3_+_95466982 | 8.34 |
ENSMUST00000090797.11
ENSMUST00000171191.6 ENSMUST00000029754.13 ENSMUST00000107154.4 |
Hormad1
|
HORMA domain containing 1 |
| chr15_-_36140539 | 8.27 |
ENSMUST00000172831.8
|
Rgs22
|
regulator of G-protein signalling 22 |
| chr17_-_40519480 | 8.26 |
ENSMUST00000033585.7
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr12_+_35034747 | 8.05 |
ENSMUST00000134550.3
|
Prps1l1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr18_+_14839202 | 7.89 |
ENSMUST00000040860.3
ENSMUST00000234680.2 |
Psma8
|
proteasome subunit alpha 8 |
| chr6_-_122778598 | 7.84 |
ENSMUST00000165884.8
|
Slc2a3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr7_-_4815542 | 7.81 |
ENSMUST00000079496.9
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr1_+_58035130 | 7.80 |
ENSMUST00000027202.9
|
Sgo2a
|
shugoshin 2A |
| chr8_+_55053809 | 7.78 |
ENSMUST00000033917.7
|
Spata4
|
spermatogenesis associated 4 |
| chr4_-_107541421 | 7.73 |
ENSMUST00000069271.5
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr2_-_125993887 | 7.55 |
ENSMUST00000110448.3
ENSMUST00000110446.9 |
Fam227b
|
family with sequence similarity 227, member B |
| chr8_-_43594523 | 7.44 |
ENSMUST00000059692.4
|
Triml1
|
tripartite motif family-like 1 |
| chr18_+_34757666 | 7.36 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr15_-_54953819 | 7.25 |
ENSMUST00000110231.2
ENSMUST00000023059.13 |
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr5_-_116427003 | 7.17 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr3_+_90173813 | 7.16 |
ENSMUST00000098914.10
|
Dennd4b
|
DENN/MADD domain containing 4B |
| chr1_-_39760150 | 7.12 |
ENSMUST00000151913.3
|
Rfx8
|
regulatory factor X 8 |
| chr9_+_48406706 | 6.98 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr11_+_106167541 | 6.88 |
ENSMUST00000044462.4
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr17_+_28426752 | 6.87 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr2_+_150940165 | 6.86 |
ENSMUST00000109888.2
|
Gm14147
|
predicted gene 14147 |
| chr4_+_100336003 | 6.83 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr9_+_108269992 | 6.70 |
ENSMUST00000192995.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr8_+_106140246 | 6.57 |
ENSMUST00000213547.2
ENSMUST00000109355.9 ENSMUST00000216765.2 |
Lrrc36
|
leucine rich repeat containing 36 |
| chr16_-_18884431 | 6.55 |
ENSMUST00000200235.2
|
Iglc3
|
immunoglobulin lambda constant 3 |
| chr2_+_84669739 | 6.54 |
ENSMUST00000146816.8
ENSMUST00000028469.14 |
Slc43a1
|
solute carrier family 43, member 1 |
| chr11_-_61233647 | 6.50 |
ENSMUST00000134423.3
ENSMUST00000093029.9 |
Slc47a2
|
solute carrier family 47, member 2 |
| chr6_+_113508636 | 6.36 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr11_+_121721058 | 6.25 |
ENSMUST00000036690.7
|
Ptchd3
|
patched domain containing 3 |
| chr11_+_3282424 | 6.19 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr1_+_181404124 | 6.14 |
ENSMUST00000208001.2
|
Dnah14
|
dynein, axonemal, heavy chain 14 |
| chr7_-_43885522 | 5.98 |
ENSMUST00000206686.2
ENSMUST00000037220.5 |
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr2_-_150934270 | 5.94 |
ENSMUST00000109890.2
|
Gm14151
|
predicted gene 14151 |
| chr7_+_106808645 | 5.92 |
ENSMUST00000098135.2
|
Rbmxl2
|
RNA binding motif protein, X-linked-like 2 |
| chr10_+_75790348 | 5.89 |
ENSMUST00000099577.4
|
Gm5134
|
predicted gene 5134 |
| chr13_+_22165687 | 5.87 |
ENSMUST00000017126.6
|
Pom121l2
|
POM121 membrane glycoprotein-like 2 (rat) |
| chr1_-_171480441 | 5.84 |
ENSMUST00000192195.6
|
Slamf7
|
SLAM family member 7 |
| chrX_+_54272236 | 5.84 |
ENSMUST00000088740.5
|
Slxl1
|
Slx-like 1 |
| chr9_+_65797519 | 5.81 |
ENSMUST00000045802.7
|
Pclaf
|
PCNA clamp associated factor |
| chr5_-_121511476 | 5.79 |
ENSMUST00000202064.2
|
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr9_-_119019450 | 5.73 |
ENSMUST00000093775.12
|
Slc22a14
|
solute carrier family 22 (organic cation transporter), member 14 |
| chr1_-_53824373 | 5.68 |
ENSMUST00000027263.14
|
Stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr18_+_52464577 | 5.65 |
ENSMUST00000025388.7
|
Ftmt
|
ferritin mitochondrial |
| chr9_-_37525009 | 5.55 |
ENSMUST00000002013.11
|
Spa17
|
sperm autoantigenic protein 17 |
| chr5_+_107479023 | 5.54 |
ENSMUST00000031215.15
ENSMUST00000112677.10 |
Brdt
|
bromodomain, testis-specific |
| chr14_-_122153185 | 5.45 |
ENSMUST00000055475.9
|
Gpr18
|
G protein-coupled receptor 18 |
| chr18_+_34758062 | 5.42 |
ENSMUST00000166044.3
|
Kif20a
|
kinesin family member 20A |
| chr7_-_97387145 | 5.30 |
ENSMUST00000084986.8
|
Aqp11
|
aquaporin 11 |
| chr5_+_104447037 | 5.29 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr9_-_37524866 | 5.27 |
ENSMUST00000214786.2
|
Spa17
|
sperm autoantigenic protein 17 |
| chr11_+_115331365 | 5.25 |
ENSMUST00000093914.5
|
Trim80
|
tripartite motif-containing 80 |
| chrX_+_9715942 | 5.25 |
ENSMUST00000057113.3
|
H2al3
|
H2A histone family member L3 |
| chrX_+_53533175 | 5.21 |
ENSMUST00000180150.8
|
Gm16405
|
predicted gene 16405 |
| chrX_+_53624178 | 5.21 |
ENSMUST00000178145.8
|
Gm16430
|
predicted gene 16430 |
| chr6_+_41165156 | 5.16 |
ENSMUST00000103277.2
|
Trbv20
|
T cell receptor beta, variable 20 |
| chr9_+_108367801 | 5.14 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr5_+_34817704 | 5.09 |
ENSMUST00000074651.11
ENSMUST00000001112.14 |
Grk4
|
G protein-coupled receptor kinase 4 |
| chr19_-_20931566 | 4.96 |
ENSMUST00000039500.4
|
Tmc1
|
transmembrane channel-like gene family 1 |
| chr7_-_35236962 | 4.92 |
ENSMUST00000193633.6
ENSMUST00000187190.7 ENSMUST00000205407.2 ENSMUST00000127472.3 |
Tdrd12
|
tudor domain containing 12 |
| chr7_+_12246415 | 4.89 |
ENSMUST00000032541.5
|
2900092C05Rik
|
RIKEN cDNA 2900092C05 gene |
| chr8_+_96038143 | 4.87 |
ENSMUST00000161029.9
|
Tepp
|
testis, prostate and placenta expressed |
| chr19_-_23251179 | 4.86 |
ENSMUST00000087556.7
ENSMUST00000223934.2 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr9_-_35481689 | 4.81 |
ENSMUST00000115110.5
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr6_-_47571901 | 4.80 |
ENSMUST00000081721.13
ENSMUST00000114618.8 ENSMUST00000114616.8 |
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr17_+_36152383 | 4.74 |
ENSMUST00000082337.13
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr8_+_104977493 | 4.74 |
ENSMUST00000034342.13
ENSMUST00000212433.2 ENSMUST00000211809.2 |
Cklf
|
chemokine-like factor |
| chr8_+_70686836 | 4.73 |
ENSMUST00000164403.8
ENSMUST00000093458.11 |
Sugp2
|
SURP and G patch domain containing 2 |
| chr8_-_3767547 | 4.72 |
ENSMUST00000058040.7
|
Clec4g
|
C-type lectin domain family 4, member g |
| chr9_-_20553576 | 4.68 |
ENSMUST00000155301.8
|
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr17_-_25946370 | 4.65 |
ENSMUST00000170070.3
ENSMUST00000048054.14 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr12_+_76450941 | 4.63 |
ENSMUST00000080449.7
|
Hspa2
|
heat shock protein 2 |
| chr11_+_29323618 | 4.63 |
ENSMUST00000040182.13
ENSMUST00000109477.2 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr8_+_96038224 | 4.63 |
ENSMUST00000098480.9
ENSMUST00000212056.2 |
Tepp
|
testis, prostate and placenta expressed |
| chr9_+_108368032 | 4.60 |
ENSMUST00000166103.9
ENSMUST00000085044.14 ENSMUST00000193678.6 ENSMUST00000178075.8 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr19_+_47853981 | 4.56 |
ENSMUST00000120645.8
|
Gsto2
|
glutathione S-transferase omega 2 |
| chrX_-_52759798 | 4.55 |
ENSMUST00000067940.3
|
1700013H16Rik
|
RIKEN cDNA 1700013H16 gene |
| chr15_+_75488524 | 4.53 |
ENSMUST00000161785.8
ENSMUST00000054555.10 ENSMUST00000161752.2 |
Zfp41
|
zinc finger protein 41 |
| chr15_-_59888446 | 4.52 |
ENSMUST00000096421.4
|
4933412E24Rik
|
RIKEN cDNA 4933412E24 gene |
| chr3_-_90150393 | 4.47 |
ENSMUST00000107369.2
|
Creb3l4
|
cAMP responsive element binding protein 3-like 4 |
| chr5_-_24235295 | 4.42 |
ENSMUST00000101513.9
|
Fam126a
|
family with sequence similarity 126, member A |
| chr1_-_88205233 | 4.39 |
ENSMUST00000065420.12
ENSMUST00000054674.15 |
Hjurp
|
Holliday junction recognition protein |
| chr11_-_46280336 | 4.38 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr2_+_13578738 | 4.35 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr19_+_34169629 | 4.33 |
ENSMUST00000239240.2
ENSMUST00000054956.15 |
Stambpl1
|
STAM binding protein like 1 |
| chr13_-_23735822 | 4.29 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr3_+_104545974 | 4.23 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr19_-_5779648 | 4.22 |
ENSMUST00000116558.3
ENSMUST00000099955.4 ENSMUST00000161368.2 |
Fam89b
|
family with sequence similarity 89, member B |
| chr5_-_113862810 | 4.19 |
ENSMUST00000047891.6
ENSMUST00000162388.8 |
1700069L16Rik
|
RIKEN cDNA 1700069L16 gene |
| chr12_-_112792971 | 4.17 |
ENSMUST00000062092.7
ENSMUST00000220899.2 |
Cdca4
|
cell division cycle associated 4 |
| chr12_+_51640097 | 4.16 |
ENSMUST00000164782.10
ENSMUST00000085412.7 |
Coch
|
cochlin |
| chr3_-_107240989 | 4.09 |
ENSMUST00000061772.11
|
Rbm15
|
RNA binding motif protein 15 |
| chr7_+_19311212 | 4.07 |
ENSMUST00000108453.2
|
Zfp296
|
zinc finger protein 296 |
| chr13_+_104365432 | 4.05 |
ENSMUST00000070761.10
ENSMUST00000225557.2 |
Cenpk
|
centromere protein K |
| chr9_-_119019428 | 3.97 |
ENSMUST00000127794.2
|
Slc22a14
|
solute carrier family 22 (organic cation transporter), member 14 |
| chr7_-_24459736 | 3.97 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr11_-_106889291 | 3.95 |
ENSMUST00000124541.8
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr11_-_83177548 | 3.95 |
ENSMUST00000163961.3
|
Slfn14
|
schlafen 14 |
| chr5_-_123662175 | 3.95 |
ENSMUST00000200247.5
ENSMUST00000111586.8 ENSMUST00000031385.7 ENSMUST00000145152.8 ENSMUST00000111587.10 |
Diablo
|
diablo, IAP-binding mitochondrial protein |
| chr11_-_98620200 | 3.93 |
ENSMUST00000126565.2
ENSMUST00000100500.9 ENSMUST00000017354.13 |
Med24
|
mediator complex subunit 24 |
| chr16_-_95993420 | 3.90 |
ENSMUST00000113804.8
ENSMUST00000054855.14 |
Lca5l
|
Leber congenital amaurosis 5-like |
| chr5_+_76736514 | 3.87 |
ENSMUST00000121979.8
|
Cep135
|
centrosomal protein 135 |
| chr1_-_156301821 | 3.87 |
ENSMUST00000188027.2
ENSMUST00000187507.7 ENSMUST00000189661.7 |
Soat1
|
sterol O-acyltransferase 1 |
| chr17_+_36152559 | 3.86 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr14_+_67953584 | 3.84 |
ENSMUST00000145542.8
ENSMUST00000125212.2 |
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr13_+_22165363 | 3.82 |
ENSMUST00000117882.2
|
Pom121l2
|
POM121 membrane glycoprotein-like 2 (rat) |
| chr4_-_119217079 | 3.82 |
ENSMUST00000143494.3
ENSMUST00000154606.9 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr13_+_21363602 | 3.81 |
ENSMUST00000222544.2
|
Trim27
|
tripartite motif-containing 27 |
| chr2_+_150751475 | 3.77 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr1_-_53824307 | 3.76 |
ENSMUST00000185920.2
|
Stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr13_+_21364069 | 3.73 |
ENSMUST00000021761.13
|
Trim27
|
tripartite motif-containing 27 |
| chr16_-_3821614 | 3.72 |
ENSMUST00000171658.2
ENSMUST00000171762.2 |
Slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr12_+_76451177 | 3.67 |
ENSMUST00000219555.2
|
Hspa2
|
heat shock protein 2 |
| chr11_-_46280298 | 3.67 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr18_+_34757687 | 3.67 |
ENSMUST00000237407.2
|
Kif20a
|
kinesin family member 20A |
| chrX_+_106193060 | 3.65 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr14_+_67953547 | 3.57 |
ENSMUST00000078053.13
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr9_+_99125420 | 3.55 |
ENSMUST00000185799.7
ENSMUST00000093795.10 ENSMUST00000190715.7 ENSMUST00000191335.7 ENSMUST00000190078.7 |
Cep70
|
centrosomal protein 70 |
| chr2_-_75812311 | 3.53 |
ENSMUST00000099994.5
|
Ttc30a1
|
tetratricopeptide repeat domain 30A1 |
| chr9_-_45895635 | 3.52 |
ENSMUST00000215427.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr19_+_46385321 | 3.52 |
ENSMUST00000039922.13
ENSMUST00000111867.9 ENSMUST00000120778.8 |
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr5_+_29400981 | 3.52 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr8_+_106434901 | 3.51 |
ENSMUST00000013302.7
ENSMUST00000211852.2 |
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr11_+_93776965 | 3.50 |
ENSMUST00000063718.11
ENSMUST00000107854.9 |
Mbtd1
|
mbt domain containing 1 |
| chr10_+_84938452 | 3.49 |
ENSMUST00000095383.6
|
Tmem263
|
transmembrane protein 263 |
| chr16_+_32219324 | 3.48 |
ENSMUST00000115149.3
|
Tm4sf19
|
transmembrane 4 L six family member 19 |
| chrX_-_47966588 | 3.45 |
ENSMUST00000114928.2
|
Fsip2l
|
fibrous sheath-interacting protein 2-like |
| chr2_-_32602246 | 3.44 |
ENSMUST00000155205.2
ENSMUST00000120105.8 |
Cdk9
|
cyclin-dependent kinase 9 (CDC2-related kinase) |
| chr11_+_106642052 | 3.42 |
ENSMUST00000147326.9
ENSMUST00000182896.8 ENSMUST00000182908.8 ENSMUST00000086353.11 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr7_+_25327028 | 3.37 |
ENSMUST00000076034.8
|
B3gnt8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr6_-_113577606 | 3.37 |
ENSMUST00000035870.5
|
Fancd2os
|
Fancd2 opposite strand |
| chrX_-_19347558 | 3.35 |
ENSMUST00000089246.6
|
Tex13c3
|
TEX13 family member C3 |
| chr12_-_69245191 | 3.33 |
ENSMUST00000021356.6
|
Dnaaf2
|
dynein, axonemal assembly factor 2 |
| chr11_+_78237492 | 3.32 |
ENSMUST00000100755.4
|
Unc119
|
unc-119 lipid binding chaperone |
| chr7_+_29979512 | 3.31 |
ENSMUST00000108187.8
ENSMUST00000014072.6 |
1700020L13Rik
|
RIKEN cDNA 1700020L13 gene |
| chr8_+_46616688 | 3.26 |
ENSMUST00000034048.13
ENSMUST00000145229.2 |
Cfap97
|
cilia and flagella associated protein 97 |
| chr13_+_23940964 | 3.26 |
ENSMUST00000102965.4
|
H4c2
|
H4 clustered histone 2 |
| chr8_+_46616770 | 3.25 |
ENSMUST00000110376.8
|
Cfap97
|
cilia and flagella associated protein 97 |
| chr12_-_112964279 | 3.25 |
ENSMUST00000011302.9
|
Brf1
|
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr11_-_106890307 | 3.21 |
ENSMUST00000018506.13
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr5_+_145077172 | 3.20 |
ENSMUST00000162594.8
ENSMUST00000162308.8 ENSMUST00000159018.8 ENSMUST00000160075.2 |
Bud31
|
BUD31 homolog |
| chr9_-_44144772 | 3.20 |
ENSMUST00000205755.2
|
Cbl
|
Casitas B-lineage lymphoma |
| chr14_+_30723340 | 3.18 |
ENSMUST00000168584.9
ENSMUST00000226378.2 |
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr14_+_44340111 | 3.17 |
ENSMUST00000074839.7
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chrY_+_1010543 | 3.14 |
ENSMUST00000091197.4
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr15_-_44291699 | 3.13 |
ENSMUST00000038719.8
|
Nudcd1
|
NudC domain containing 1 |
| chr18_-_46658957 | 3.13 |
ENSMUST00000036226.6
|
Fem1c
|
fem 1 homolog c |
| chr7_+_140871901 | 3.12 |
ENSMUST00000026569.6
|
Drd4
|
dopamine receptor D4 |
| chr4_+_140633646 | 3.08 |
ENSMUST00000030765.7
|
Padi2
|
peptidyl arginine deiminase, type II |
| chr4_-_62389098 | 3.06 |
ENSMUST00000135811.2
ENSMUST00000120095.8 ENSMUST00000030087.14 ENSMUST00000107452.8 ENSMUST00000155522.8 |
Wdr31
|
WD repeat domain 31 |
| chr7_-_30298287 | 3.05 |
ENSMUST00000108150.2
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr1_-_191307648 | 3.03 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase |
| chr14_+_30723371 | 3.02 |
ENSMUST00000022476.9
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr11_-_62430001 | 3.01 |
ENSMUST00000018653.8
|
Cenpv
|
centromere protein V |
| chr6_+_47021979 | 2.99 |
ENSMUST00000150737.2
|
Cntnap2
|
contactin associated protein-like 2 |
| chr4_+_62443606 | 2.99 |
ENSMUST00000062145.2
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr5_+_35106778 | 2.98 |
ENSMUST00000030984.14
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr2_+_157579321 | 2.96 |
ENSMUST00000029178.7
|
Ctnnbl1
|
catenin, beta like 1 |
| chr14_+_55983136 | 2.94 |
ENSMUST00000019441.9
|
Nop9
|
NOP9 nucleolar protein |
| chrX_-_8327965 | 2.94 |
ENSMUST00000103000.9
ENSMUST00000023931.4 |
Ssxb2
|
synovial sarcoma, X member B2 |
| chr5_-_122510292 | 2.90 |
ENSMUST00000031419.6
|
Fam216a
|
family with sequence similarity 216, member A |
| chr18_+_34891941 | 2.85 |
ENSMUST00000049281.12
|
Fam53c
|
family with sequence similarity 53, member C |
| chr3_-_130524024 | 2.85 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr15_-_79212400 | 2.84 |
ENSMUST00000173163.8
ENSMUST00000047816.15 ENSMUST00000172403.9 ENSMUST00000173632.8 |
Pla2g6
|
phospholipase A2, group VI |
| chr7_-_97387429 | 2.80 |
ENSMUST00000206389.2
|
Aqp11
|
aquaporin 11 |
| chr17_-_46513499 | 2.80 |
ENSMUST00000024749.9
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chr18_+_11790888 | 2.77 |
ENSMUST00000234499.2
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr14_+_53903370 | 2.75 |
ENSMUST00000181768.3
|
Trav3-3
|
T cell receptor alpha variable 3-3 |
| chr13_+_49806772 | 2.74 |
ENSMUST00000223264.2
ENSMUST00000221142.2 ENSMUST00000222333.2 ENSMUST00000021824.8 |
Nol8
|
nucleolar protein 8 |
| chr3_-_95902949 | 2.73 |
ENSMUST00000123006.8
ENSMUST00000130043.8 |
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr10_+_111134525 | 2.72 |
ENSMUST00000040454.5
|
Bbs10
|
Bardet-Biedl syndrome 10 (human) |
| chr7_+_55417967 | 2.68 |
ENSMUST00000060416.15
ENSMUST00000094360.13 ENSMUST00000165045.9 ENSMUST00000173835.2 |
Siglech
|
sialic acid binding Ig-like lectin H |
| chr14_+_54183465 | 2.66 |
ENSMUST00000197130.5
ENSMUST00000103677.3 |
Trdv2-1
|
T cell receptor delta variable 2-1 |
| chr12_+_103277234 | 2.65 |
ENSMUST00000191218.7
|
Fam181a
|
family with sequence similarity 181, member A |
| chr13_-_63036096 | 2.61 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr8_+_104977551 | 2.61 |
ENSMUST00000098464.6
|
Cklf
|
chemokine-like factor |
| chr17_+_15198730 | 2.59 |
ENSMUST00000061688.11
|
9030025P20Rik
|
RIKEN cDNA 9030025P20 gene |
| chr3_-_95903313 | 2.58 |
ENSMUST00000015889.10
|
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 3.2 | 9.7 | GO:0051878 | lateral element assembly(GO:0051878) |
| 2.8 | 8.3 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 2.5 | 12.3 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 2.1 | 8.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 2.0 | 8.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 2.0 | 14.0 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) |
| 1.8 | 5.4 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 1.6 | 4.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 1.6 | 7.8 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 1.6 | 7.8 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.4 | 7.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.4 | 4.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.4 | 4.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 1.3 | 3.9 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 1.3 | 7.5 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 1.2 | 3.7 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) positive regulation of t-circle formation(GO:1904431) |
| 1.2 | 7.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 1.2 | 6.9 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.0 | 9.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.9 | 6.0 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.8 | 3.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.8 | 3.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.8 | 5.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.8 | 2.4 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.8 | 3.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.8 | 4.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.8 | 3.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.7 | 16.5 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.7 | 32.9 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.7 | 2.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.7 | 18.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.7 | 2.6 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.6 | 9.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.6 | 1.8 | GO:0002481 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.6 | 3.0 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.6 | 12.8 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.6 | 4.7 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.6 | 3.4 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.6 | 5.7 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.6 | 6.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.6 | 1.7 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.5 | 4.4 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.5 | 5.9 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.5 | 4.6 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.5 | 1.0 | GO:0072708 | response to sorbitol(GO:0072708) |
| 0.5 | 5.0 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.5 | 2.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.5 | 9.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.5 | 10.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.5 | 2.8 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.5 | 2.8 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.5 | 1.4 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.5 | 4.6 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.5 | 1.4 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.4 | 17.8 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.4 | 3.9 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 7.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.4 | 3.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.4 | 7.8 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.4 | 2.0 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.4 | 1.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 4.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.4 | 3.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.4 | 4.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.3 | 2.8 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.3 | 3.8 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.3 | 18.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 1.0 | GO:0070318 | enucleate erythrocyte development(GO:0048822) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.3 | 4.0 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.3 | 4.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 0.9 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.3 | 5.9 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.3 | 2.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.3 | 0.6 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 8.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.3 | 1.5 | GO:0035822 | meiotic gene conversion(GO:0006311) male meiosis chromosome segregation(GO:0007060) gene conversion(GO:0035822) |
| 0.3 | 0.9 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.3 | 5.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.3 | 0.5 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 0.2 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.2 | 4.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 2.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.2 | 1.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 5.0 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.2 | 2.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.9 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 3.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 5.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 3.2 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.2 | 1.5 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 1.5 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.2 | 10.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 3.7 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 4.7 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.2 | 0.6 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.2 | 1.9 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 1.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.2 | 1.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.2 | 1.5 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.2 | 4.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.2 | 2.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 1.8 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 10.0 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.2 | 1.0 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.2 | 2.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 5.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.2 | 1.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.4 | GO:2000407 | T cell extravasation(GO:0072683) regulation of T cell extravasation(GO:2000407) positive regulation of T cell extravasation(GO:2000409) |
| 0.1 | 0.9 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.7 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 1.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 1.3 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 0.6 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 7.3 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 6.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 1.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 9.9 | GO:0070228 | regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.1 | 7.0 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.1 | 3.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 4.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 6.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 0.8 | GO:0071543 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 7.1 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 1.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 9.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 2.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 13.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.5 | GO:0021763 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.7 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 5.8 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 1.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 3.1 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 2.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 1.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 2.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 3.0 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.1 | 2.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 1.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 5.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.3 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 0.7 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 3.2 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.1 | 2.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.4 | GO:0050000 | chromosome localization(GO:0050000) establishment of chromosome localization(GO:0051303) |
| 0.1 | 5.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 8.6 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 7.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.9 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 4.0 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 13.2 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 1.1 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 2.9 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 3.0 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 8.1 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 8.2 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 12.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.2 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 4.0 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 1.9 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 4.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.6 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 4.1 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 2.0 | GO:0042254 | ribosome biogenesis(GO:0042254) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 34.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.9 | 9.7 | GO:0000802 | transverse filament(GO:0000802) |
| 1.6 | 9.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.5 | 14.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 1.3 | 7.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.3 | 3.8 | GO:0000811 | GINS complex(GO:0000811) |
| 1.2 | 11.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.1 | 7.8 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.9 | 8.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.9 | 6.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.8 | 8.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.8 | 9.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 7.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.7 | 3.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.7 | 4.9 | GO:1990923 | PET complex(GO:1990923) |
| 0.5 | 5.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.5 | 13.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.5 | 2.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.5 | 13.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.4 | 12.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.4 | 2.6 | GO:0070449 | elongin complex(GO:0070449) |
| 0.4 | 5.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.4 | 1.2 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.4 | 4.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.4 | 12.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 2.4 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.3 | 2.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 1.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 3.4 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.3 | 22.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 1.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.3 | 1.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 3.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.3 | 7.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 4.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 7.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 6.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 3.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 14.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 51.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.2 | 6.9 | GO:0045120 | pronucleus(GO:0045120) |
| 0.2 | 6.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 5.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 0.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 3.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 3.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 7.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 8.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.4 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 0.6 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 3.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 7.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 2.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.0 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 3.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 17.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 8.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.2 | GO:0042272 | nucleocytoplasmic shuttling complex(GO:0031074) nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 3.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 2.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 3.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.8 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 0.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.2 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.1 | 7.4 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 5.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 5.0 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 3.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 33.7 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 4.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.1 | GO:0032994 | protein-lipid complex(GO:0032994) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 6.9 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 3.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 3.5 | 10.4 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.7 | 10.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.7 | 8.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 1.6 | 7.8 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 1.5 | 6.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.1 | 3.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 1.1 | 11.9 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.0 | 7.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.7 | 9.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.7 | 3.7 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.7 | 5.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.7 | 4.6 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.6 | 3.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.6 | 4.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.6 | 3.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.6 | 2.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.6 | 12.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.5 | 3.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.5 | 5.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 4.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.5 | 3.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.5 | 5.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 4.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.5 | 18.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.5 | 1.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.5 | 8.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 1.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 5.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.4 | 1.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 1.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 9.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 1.0 | GO:0001007 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) |
| 0.3 | 1.0 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.3 | 4.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 8.3 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.3 | 7.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 3.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 0.9 | GO:0001571 | non-tyrosine kinase fibroblast growth factor receptor activity(GO:0001571) |
| 0.3 | 2.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.3 | 4.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 31.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 9.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 1.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.3 | 2.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 3.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 8.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 1.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 0.9 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.2 | 4.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 3.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 0.6 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.2 | 5.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 2.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 0.9 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 2.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 2.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 2.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.2 | 8.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 4.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 0.8 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 12.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 4.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 3.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 3.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 7.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.7 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 6.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 10.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 3.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.8 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 4.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 13.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.3 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.1 | 4.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 3.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 5.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.6 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 3.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 4.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 3.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 4.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.1 | 12.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 5.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 1.5 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 6.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.0 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 2.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 4.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 4.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 6.4 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 3.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 3.5 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 19.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.9 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 2.1 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 2.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 2.1 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 1.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 7.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 3.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 30.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.5 | 19.7 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 3.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.3 | 8.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 11.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 9.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 9.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 7.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 14.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 6.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 4.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 3.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 5.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.8 | 25.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.7 | 9.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.6 | 10.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 10.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.6 | 10.9 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.6 | 4.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.5 | 12.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.5 | 7.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 18.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.4 | 10.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 43.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 9.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 3.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 3.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 1.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 5.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 4.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 7.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 3.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 8.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 2.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 6.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.8 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 0.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 6.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 14.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 4.9 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 7.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 3.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 2.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.8 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 2.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 6.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |