Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
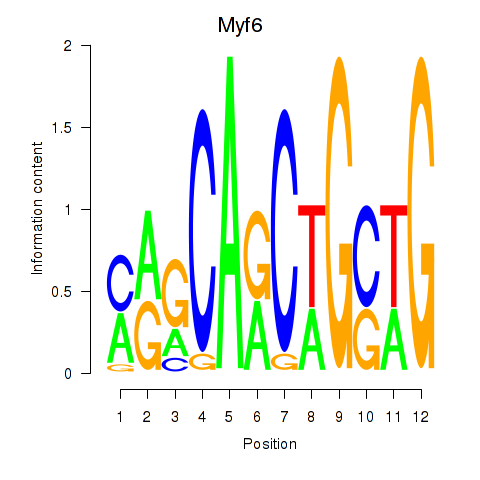
Results for Myf6
Z-value: 2.39
Transcription factors associated with Myf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myf6
|
ENSMUSG00000035923.5 | Myf6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myf6 | mm39_v1_chr10_-_107330580_107330603 | -0.05 | 7.0e-01 | Click! |
Activity profile of Myf6 motif
Sorted Z-values of Myf6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Myf6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_9821021 | 34.44 |
ENSMUST00000208933.2
ENSMUST00000110969.5 |
Fam155a
|
family with sequence similarity 155, member A |
| chr18_+_13088878 | 21.55 |
ENSMUST00000234763.2
|
Impact
|
impact, RWD domain protein |
| chrX_+_72546680 | 19.28 |
ENSMUST00000033744.12
ENSMUST00000088429.8 ENSMUST00000114479.2 |
Atp2b3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr9_-_112015007 | 19.19 |
ENSMUST00000070218.12
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr2_+_83642910 | 18.16 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr1_+_181180183 | 16.97 |
ENSMUST00000161880.8
ENSMUST00000027795.14 |
Cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr6_+_118043307 | 15.47 |
ENSMUST00000203804.3
ENSMUST00000203482.2 |
Rasgef1a
|
RasGEF domain family, member 1A |
| chr4_+_129878890 | 14.68 |
ENSMUST00000106017.8
ENSMUST00000121049.8 |
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr12_+_105420089 | 14.59 |
ENSMUST00000178224.2
|
D430019H16Rik
|
RIKEN cDNA D430019H16 gene |
| chr6_+_107506678 | 14.55 |
ENSMUST00000049285.10
|
Lrrn1
|
leucine rich repeat protein 1, neuronal |
| chr11_+_78213791 | 14.42 |
ENSMUST00000017534.15
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr4_-_60777462 | 14.37 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr16_+_41353360 | 14.20 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr4_-_61259997 | 14.15 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr3_-_116047148 | 14.08 |
ENSMUST00000090473.7
|
Gpr88
|
G-protein coupled receptor 88 |
| chr16_-_28571820 | 13.94 |
ENSMUST00000232352.2
|
Fgf12
|
fibroblast growth factor 12 |
| chr12_+_108300599 | 13.75 |
ENSMUST00000021684.6
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr6_-_126717590 | 13.63 |
ENSMUST00000185333.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr4_-_60377932 | 13.24 |
ENSMUST00000107506.9
ENSMUST00000122381.8 ENSMUST00000118759.8 ENSMUST00000132829.3 |
Mup9
|
major urinary protein 9 |
| chr4_-_60070411 | 13.21 |
ENSMUST00000079697.10
ENSMUST00000125282.2 ENSMUST00000166098.8 |
Mup7
|
major urinary protein 7 |
| chr4_-_60618357 | 13.01 |
ENSMUST00000084544.5
ENSMUST00000098046.10 |
Mup11
|
major urinary protein 11 |
| chr4_-_60222580 | 12.95 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chrX_+_158242121 | 12.85 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr16_+_91066602 | 12.80 |
ENSMUST00000056882.7
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr4_-_60139857 | 12.80 |
ENSMUST00000107490.5
ENSMUST00000074700.9 |
Mup2
|
major urinary protein 2 |
| chr6_-_126717114 | 12.80 |
ENSMUST00000112242.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr4_-_61592331 | 12.63 |
ENSMUST00000098040.4
|
Mup18
|
major urinary protein 18 |
| chr4_-_61259801 | 12.54 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr10_+_7465555 | 12.25 |
ENSMUST00000134346.8
ENSMUST00000019931.12 ENSMUST00000130590.8 |
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr17_+_35455532 | 12.24 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr4_-_60457902 | 12.21 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr12_-_72455708 | 11.51 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr14_-_24054927 | 11.49 |
ENSMUST00000145596.3
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_-_148574353 | 11.44 |
ENSMUST00000028926.13
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta |
| chr4_-_60697274 | 11.42 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr7_-_127410096 | 11.27 |
ENSMUST00000156135.3
|
Stx1b
|
syntaxin 1B |
| chr7_+_87233554 | 10.81 |
ENSMUST00000125009.9
|
Grm5
|
glutamate receptor, metabotropic 5 |
| chr4_-_149211145 | 10.74 |
ENSMUST00000030815.3
|
Cort
|
cortistatin |
| chr6_-_36787096 | 10.53 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr8_+_94879235 | 10.52 |
ENSMUST00000034211.10
ENSMUST00000211930.2 ENSMUST00000211915.2 |
Mt3
|
metallothionein 3 |
| chr7_+_3352159 | 10.34 |
ENSMUST00000172109.4
|
Prkcg
|
protein kinase C, gamma |
| chr4_-_61437704 | 10.30 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr12_-_119202527 | 10.29 |
ENSMUST00000026360.9
|
Itgb8
|
integrin beta 8 |
| chr10_+_90412570 | 10.24 |
ENSMUST00000182430.8
ENSMUST00000182960.8 ENSMUST00000182045.2 ENSMUST00000182083.2 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_-_77956635 | 10.23 |
ENSMUST00000161846.8
ENSMUST00000160894.8 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr7_+_98916635 | 10.22 |
ENSMUST00000122101.8
ENSMUST00000068973.11 ENSMUST00000207883.2 |
Map6
|
microtubule-associated protein 6 |
| chr10_+_90412638 | 10.14 |
ENSMUST00000183136.8
ENSMUST00000182595.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_+_29087658 | 10.00 |
ENSMUST00000213489.2
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr1_-_75240551 | 9.94 |
ENSMUST00000186178.7
ENSMUST00000189769.7 ENSMUST00000027404.12 |
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr7_+_3352019 | 9.84 |
ENSMUST00000100301.11
|
Prkcg
|
protein kinase C, gamma |
| chr2_-_151474391 | 9.64 |
ENSMUST00000137936.2
ENSMUST00000146172.8 ENSMUST00000094456.10 ENSMUST00000148755.8 ENSMUST00000109875.8 ENSMUST00000028951.14 ENSMUST00000109877.10 |
Snph
|
syntaphilin |
| chr12_+_16703383 | 9.55 |
ENSMUST00000221596.2
ENSMUST00000111064.3 ENSMUST00000220892.2 |
Ntsr2
|
neurotensin receptor 2 |
| chr7_+_121888520 | 9.51 |
ENSMUST00000064989.12
ENSMUST00000064921.5 |
Prkcb
|
protein kinase C, beta |
| chr2_-_25209107 | 9.45 |
ENSMUST00000114318.10
ENSMUST00000114310.10 ENSMUST00000114308.10 ENSMUST00000114317.10 ENSMUST00000028335.13 ENSMUST00000114314.10 ENSMUST00000114307.8 |
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chrX_-_102230225 | 9.34 |
ENSMUST00000121720.2
|
Nap1l2
|
nucleosome assembly protein 1-like 2 |
| chr5_-_128897086 | 9.33 |
ENSMUST00000198941.5
ENSMUST00000199537.5 |
Rimbp2
|
RIMS binding protein 2 |
| chrX_-_121307036 | 9.27 |
ENSMUST00000079490.6
|
Nap1l3
|
nucleosome assembly protein 1-like 3 |
| chr8_+_70768409 | 9.27 |
ENSMUST00000165819.9
ENSMUST00000140239.4 |
Cers1
|
ceramide synthase 1 |
| chr6_-_77956499 | 9.24 |
ENSMUST00000159626.8
ENSMUST00000075340.12 ENSMUST00000162273.2 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr7_-_141009264 | 9.13 |
ENSMUST00000164387.2
ENSMUST00000137488.2 ENSMUST00000084436.10 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr6_-_121450547 | 9.09 |
ENSMUST00000046373.8
ENSMUST00000151397.3 |
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr15_+_38940307 | 9.05 |
ENSMUST00000067072.5
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr16_+_41353212 | 8.98 |
ENSMUST00000078873.11
|
Lsamp
|
limbic system-associated membrane protein |
| chr10_+_29087602 | 8.95 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr2_+_21372338 | 8.86 |
ENSMUST00000055946.8
|
Gpr158
|
G protein-coupled receptor 158 |
| chr7_+_51528715 | 8.76 |
ENSMUST00000051912.13
|
Gas2
|
growth arrest specific 2 |
| chr8_-_70939964 | 8.58 |
ENSMUST00000045286.9
|
Tmem59l
|
transmembrane protein 59-like |
| chr12_-_70394074 | 8.56 |
ENSMUST00000223160.2
ENSMUST00000222316.2 ENSMUST00000167755.3 ENSMUST00000110520.10 ENSMUST00000110522.10 ENSMUST00000221041.2 ENSMUST00000222603.3 |
Trim9
|
tripartite motif-containing 9 |
| chr15_-_37459570 | 8.55 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chrX_+_98864627 | 8.54 |
ENSMUST00000096363.3
|
Tmem28
|
transmembrane protein 28 |
| chr4_-_15149051 | 8.51 |
ENSMUST00000041606.14
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr10_+_90412827 | 8.46 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr7_-_12732067 | 8.43 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr1_+_162466717 | 8.41 |
ENSMUST00000028020.11
|
Myoc
|
myocilin |
| chrX_-_7439082 | 8.38 |
ENSMUST00000132788.2
|
Ppp1r3f
|
protein phosphatase 1, regulatory subunit 3F |
| chr2_-_180681079 | 8.37 |
ENSMUST00000067120.14
|
Chrna4
|
cholinergic receptor, nicotinic, alpha polypeptide 4 |
| chr10_+_90412432 | 8.34 |
ENSMUST00000182786.8
ENSMUST00000182600.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr17_-_91396154 | 8.33 |
ENSMUST00000161402.10
ENSMUST00000054059.15 ENSMUST00000072671.14 ENSMUST00000174331.8 |
Nrxn1
|
neurexin I |
| chr16_-_20440005 | 8.33 |
ENSMUST00000052939.4
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chrX_-_71699740 | 8.27 |
ENSMUST00000055966.13
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr15_+_38940351 | 8.27 |
ENSMUST00000226433.2
ENSMUST00000132192.3 |
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr6_-_113172340 | 8.21 |
ENSMUST00000162280.2
|
Lhfpl4
|
lipoma HMGIC fusion partner-like protein 4 |
| chr7_-_141009346 | 8.20 |
ENSMUST00000124444.2
|
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chrX_-_134111421 | 8.11 |
ENSMUST00000033783.2
|
Tceal6
|
transcription elongation factor A (SII)-like 6 |
| chr11_+_103540391 | 8.10 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chr7_-_30826376 | 8.01 |
ENSMUST00000098548.8
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr9_+_45281483 | 7.94 |
ENSMUST00000085939.8
ENSMUST00000217381.2 |
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr7_+_57069417 | 7.78 |
ENSMUST00000085240.11
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr10_-_20424101 | 7.73 |
ENSMUST00000164195.2
|
Pde7b
|
phosphodiesterase 7B |
| chr10_-_20424069 | 7.72 |
ENSMUST00000169404.8
|
Pde7b
|
phosphodiesterase 7B |
| chr2_+_55327110 | 7.71 |
ENSMUST00000112633.3
ENSMUST00000112632.2 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr5_+_22951015 | 7.67 |
ENSMUST00000197992.2
|
Lhfpl3
|
lipoma HMGIC fusion partner-like 3 |
| chr13_-_12121831 | 7.63 |
ENSMUST00000021750.15
ENSMUST00000170156.3 ENSMUST00000220597.2 |
Ryr2
|
ryanodine receptor 2, cardiac |
| chr10_+_63222338 | 7.60 |
ENSMUST00000043317.7
|
Dnajc12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr13_+_88969591 | 7.58 |
ENSMUST00000118731.8
ENSMUST00000081769.13 |
Edil3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr11_+_69914746 | 7.54 |
ENSMUST00000124568.10
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr10_+_90412539 | 7.52 |
ENSMUST00000182284.8
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr17_+_8559539 | 7.50 |
ENSMUST00000163887.2
|
Prr18
|
proline rich 18 |
| chr14_+_66205932 | 7.46 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr10_+_7465845 | 7.43 |
ENSMUST00000135907.8
|
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr11_+_104122216 | 7.42 |
ENSMUST00000106992.10
|
Mapt
|
microtubule-associated protein tau |
| chr17_+_44112679 | 7.40 |
ENSMUST00000229744.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr17_+_74111823 | 7.39 |
ENSMUST00000024860.9
|
Ehd3
|
EH-domain containing 3 |
| chr14_-_123864471 | 7.39 |
ENSMUST00000000201.7
|
Nalcn
|
sodium leak channel, non-selective |
| chr7_+_122270599 | 7.39 |
ENSMUST00000182563.2
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr7_+_24181416 | 7.39 |
ENSMUST00000068023.8
|
Cadm4
|
cell adhesion molecule 4 |
| chr18_-_43032359 | 7.37 |
ENSMUST00000117687.8
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr9_+_110075133 | 7.34 |
ENSMUST00000199736.2
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr10_+_90412114 | 7.31 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr5_+_117552042 | 7.24 |
ENSMUST00000180430.2
|
Ksr2
|
kinase suppressor of ras 2 |
| chrX_-_143471176 | 7.23 |
ENSMUST00000040184.4
|
Trpc5
|
transient receptor potential cation channel, subfamily C, member 5 |
| chr7_-_30826184 | 7.20 |
ENSMUST00000211945.2
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr13_-_95581393 | 7.19 |
ENSMUST00000221025.2
|
Crhbp
|
corticotropin releasing hormone binding protein |
| chr2_+_118610184 | 7.17 |
ENSMUST00000063975.10
ENSMUST00000037547.9 ENSMUST00000110846.8 ENSMUST00000110843.2 |
Disp2
|
dispatched RND tramsporter family member 2 |
| chr5_-_113458563 | 7.16 |
ENSMUST00000154248.8
ENSMUST00000112325.8 ENSMUST00000048112.13 |
Sgsm1
|
small G protein signaling modulator 1 |
| chr7_-_126620378 | 7.16 |
ENSMUST00000159916.5
|
Prrt2
|
proline-rich transmembrane protein 2 |
| chr15_+_89417017 | 7.15 |
ENSMUST00000167173.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr10_+_52566616 | 7.12 |
ENSMUST00000105473.3
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr18_-_43820759 | 7.05 |
ENSMUST00000082254.8
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr11_+_104122399 | 7.04 |
ENSMUST00000132977.8
ENSMUST00000132245.8 ENSMUST00000100347.11 |
Mapt
|
microtubule-associated protein tau |
| chr16_+_7042168 | 7.04 |
ENSMUST00000231088.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr8_+_63404228 | 7.02 |
ENSMUST00000118003.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr12_+_16703709 | 6.94 |
ENSMUST00000221049.2
|
Ntsr2
|
neurotensin receptor 2 |
| chr7_+_51528788 | 6.92 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr2_-_30364219 | 6.90 |
ENSMUST00000065134.4
|
Ier5l
|
immediate early response 5-like |
| chr12_-_87037204 | 6.89 |
ENSMUST00000222543.2
|
Zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr2_-_113659360 | 6.87 |
ENSMUST00000024005.8
|
Scg5
|
secretogranin V |
| chr6_+_21215472 | 6.87 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr7_+_122270623 | 6.86 |
ENSMUST00000182095.2
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr1_+_72863641 | 6.85 |
ENSMUST00000047328.11
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr13_-_95581335 | 6.84 |
ENSMUST00000045583.9
|
Crhbp
|
corticotropin releasing hormone binding protein |
| chr13_+_88969725 | 6.81 |
ENSMUST00000043111.7
|
Edil3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr18_-_43032535 | 6.80 |
ENSMUST00000120632.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chrX_-_135104589 | 6.79 |
ENSMUST00000066819.11
|
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr6_-_37276885 | 6.78 |
ENSMUST00000101532.10
|
Dgki
|
diacylglycerol kinase, iota |
| chr2_-_122441719 | 6.77 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr11_+_71640739 | 6.76 |
ENSMUST00000150531.2
|
Wscd1
|
WSC domain containing 1 |
| chr4_+_105014536 | 6.75 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr6_-_138399896 | 6.75 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr13_+_93908138 | 6.75 |
ENSMUST00000091403.6
|
Arsb
|
arylsulfatase B |
| chr14_-_55150547 | 6.74 |
ENSMUST00000228495.3
ENSMUST00000228119.3 ENSMUST00000050772.10 ENSMUST00000231305.2 |
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr10_+_57660948 | 6.68 |
ENSMUST00000020024.12
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr16_-_67417768 | 6.63 |
ENSMUST00000114292.8
ENSMUST00000120898.8 |
Cadm2
|
cell adhesion molecule 2 |
| chrX_+_20570145 | 6.63 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr6_-_12749192 | 6.58 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr8_-_85663976 | 6.53 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr11_+_104122341 | 6.49 |
ENSMUST00000106993.10
|
Mapt
|
microtubule-associated protein tau |
| chr13_+_64309675 | 6.44 |
ENSMUST00000021929.10
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr4_-_155870321 | 6.42 |
ENSMUST00000097742.3
|
Tmem88b
|
transmembrane protein 88B |
| chr3_-_141637245 | 6.26 |
ENSMUST00000106232.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr1_-_124773767 | 6.24 |
ENSMUST00000239072.2
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr7_+_143838149 | 6.23 |
ENSMUST00000146006.3
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr18_-_43032514 | 6.21 |
ENSMUST00000236238.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_+_25318642 | 6.18 |
ENSMUST00000102919.4
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr8_-_95422851 | 6.17 |
ENSMUST00000034227.6
|
Pllp
|
plasma membrane proteolipid |
| chr1_+_75526225 | 6.10 |
ENSMUST00000154101.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr11_-_69496655 | 6.10 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr10_-_93375832 | 6.10 |
ENSMUST00000016034.3
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr1_+_146373352 | 6.09 |
ENSMUST00000132847.8
ENSMUST00000166814.8 |
Brinp3
|
bone morphogenetic protein/retinoic acid inducible neural specific 3 |
| chr2_-_180680868 | 6.09 |
ENSMUST00000108851.8
|
Chrna4
|
cholinergic receptor, nicotinic, alpha polypeptide 4 |
| chr4_+_115375461 | 6.05 |
ENSMUST00000058785.10
ENSMUST00000094886.4 |
Cyp4a10
|
cytochrome P450, family 4, subfamily a, polypeptide 10 |
| chr7_+_126549859 | 6.04 |
ENSMUST00000106333.8
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr18_+_57266123 | 6.02 |
ENSMUST00000075770.13
|
Megf10
|
multiple EGF-like-domains 10 |
| chr11_+_119833589 | 5.95 |
ENSMUST00000106231.8
ENSMUST00000075180.12 ENSMUST00000103021.10 ENSMUST00000026436.10 ENSMUST00000106233.2 |
Baiap2
|
brain-specific angiogenesis inhibitor 1-associated protein 2 |
| chr8_-_88198992 | 5.86 |
ENSMUST00000169693.2
|
Cbln1
|
cerebellin 1 precursor protein |
| chr11_+_104122291 | 5.86 |
ENSMUST00000145227.8
|
Mapt
|
microtubule-associated protein tau |
| chr7_+_26821266 | 5.85 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr9_-_43151179 | 5.83 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr2_-_25209199 | 5.80 |
ENSMUST00000114312.2
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr14_+_55728519 | 5.78 |
ENSMUST00000076236.7
|
Carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr7_+_126550009 | 5.74 |
ENSMUST00000106332.3
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chrX_+_133305529 | 5.74 |
ENSMUST00000113224.9
ENSMUST00000113226.2 |
Drp2
|
dystrophin related protein 2 |
| chr11_+_102652228 | 5.69 |
ENSMUST00000103081.10
ENSMUST00000068150.7 |
Adam11
|
a disintegrin and metallopeptidase domain 11 |
| chr10_+_57661010 | 5.68 |
ENSMUST00000165013.2
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr2_-_162502994 | 5.68 |
ENSMUST00000109442.8
ENSMUST00000109445.9 ENSMUST00000109443.8 ENSMUST00000109441.2 |
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr16_-_22475915 | 5.67 |
ENSMUST00000089925.10
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr6_-_21851827 | 5.65 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr2_-_131953359 | 5.64 |
ENSMUST00000128899.2
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr2_+_91757594 | 5.63 |
ENSMUST00000045537.4
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chr12_+_105302853 | 5.62 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr15_-_89033761 | 5.60 |
ENSMUST00000088823.5
|
Mapk11
|
mitogen-activated protein kinase 11 |
| chr13_-_65051763 | 5.56 |
ENSMUST00000091554.6
|
Cntnap3
|
contactin associated protein-like 3 |
| chr11_-_6015538 | 5.50 |
ENSMUST00000101585.10
ENSMUST00000066431.14 ENSMUST00000109815.9 ENSMUST00000109812.9 ENSMUST00000101586.3 ENSMUST00000093355.12 ENSMUST00000019133.11 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta |
| chr7_+_98916677 | 5.50 |
ENSMUST00000127492.2
|
Map6
|
microtubule-associated protein 6 |
| chr2_-_23938869 | 5.44 |
ENSMUST00000114497.2
|
Hnmt
|
histamine N-methyltransferase |
| chr4_-_66322695 | 5.42 |
ENSMUST00000084496.3
|
Astn2
|
astrotactin 2 |
| chrX_+_152615221 | 5.42 |
ENSMUST00000148708.2
ENSMUST00000123264.2 ENSMUST00000049999.9 |
Spin2c
|
spindlin family, member 2C |
| chr4_-_46991842 | 5.40 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chrX_-_135104386 | 5.40 |
ENSMUST00000151592.8
ENSMUST00000131510.2 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr16_-_74208395 | 5.39 |
ENSMUST00000227347.2
|
Robo2
|
roundabout guidance receptor 2 |
| chr10_+_127702326 | 5.38 |
ENSMUST00000092058.4
|
Rdh16f2
|
RDH16 family member 2 |
| chr16_-_60425608 | 5.35 |
ENSMUST00000068860.13
|
Epha6
|
Eph receptor A6 |
| chr2_-_119590776 | 5.34 |
ENSMUST00000082130.13
ENSMUST00000028759.13 |
Ltk
|
leukocyte tyrosine kinase |
| chr10_-_32765671 | 5.32 |
ENSMUST00000218645.2
|
Nkain2
|
Na+/K+ transporting ATPase interacting 2 |
| chr5_-_103359117 | 5.29 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr4_+_129878627 | 5.26 |
ENSMUST00000120204.8
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr7_+_4122523 | 5.26 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr8_-_88199231 | 5.21 |
ENSMUST00000034076.16
|
Cbln1
|
cerebellin 1 precursor protein |
| chr5_+_71857261 | 5.20 |
ENSMUST00000031122.9
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chrX_-_52672363 | 5.19 |
ENSMUST00000088778.5
|
Rtl8b
|
retrotransposon Gag like 8B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.5 | GO:1901561 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 5.1 | 15.2 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 5.1 | 15.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 4.8 | 14.5 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 4.7 | 52.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 4.3 | 17.3 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 4.1 | 12.4 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 3.6 | 10.8 | GO:1902938 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 3.5 | 10.5 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 3.5 | 14.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 3.4 | 41.1 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 3.4 | 20.2 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 3.3 | 13.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 3.3 | 9.9 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 3.2 | 9.6 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 3.0 | 21.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 3.0 | 3.0 | GO:0032805 | positive regulation of low-density lipoprotein particle receptor catabolic process(GO:0032805) |
| 2.9 | 5.8 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 2.8 | 13.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 2.7 | 26.8 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 2.5 | 7.6 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 2.5 | 17.7 | GO:0061743 | motor learning(GO:0061743) |
| 2.5 | 19.7 | GO:0009414 | response to water deprivation(GO:0009414) |
| 2.4 | 7.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 2.3 | 25.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 2.3 | 13.6 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 2.3 | 6.8 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 2.3 | 11.3 | GO:1904048 | negative regulation of synaptic vesicle recycling(GO:1903422) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 2.2 | 6.7 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 2.2 | 8.9 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 2.1 | 10.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 2.1 | 8.4 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 1.9 | 11.7 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.9 | 28.9 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 1.9 | 5.6 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.8 | 5.5 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 1.7 | 10.3 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 1.7 | 5.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 1.6 | 11.5 | GO:0052805 | imidazole-containing compound catabolic process(GO:0052805) |
| 1.5 | 3.0 | GO:0036301 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 1.5 | 15.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 1.5 | 8.8 | GO:0036394 | amylase secretion(GO:0036394) |
| 1.4 | 4.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.4 | 4.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.4 | 5.6 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 1.4 | 8.3 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 1.3 | 4.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 1.3 | 5.2 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 1.3 | 13.0 | GO:0046959 | habituation(GO:0046959) |
| 1.3 | 3.8 | GO:1904434 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 1.3 | 3.8 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 1.3 | 6.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.2 | 8.7 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.2 | 5.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 1.2 | 4.9 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.2 | 7.3 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.2 | 6.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 1.2 | 8.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 1.2 | 9.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 1.2 | 3.5 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 1.1 | 30.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 1.1 | 2.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 1.1 | 20.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 1.1 | 39.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 1.1 | 5.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.1 | 3.4 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 1.1 | 3.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.1 | 13.7 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.1 | 3.2 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 1.0 | 9.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 1.0 | 5.0 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 1.0 | 3.9 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.0 | 11.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.9 | 7.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.9 | 0.9 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.9 | 8.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.9 | 10.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.9 | 11.4 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.9 | 8.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.8 | 2.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.8 | 2.4 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.8 | 13.7 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.8 | 9.5 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.8 | 8.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.8 | 7.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.8 | 35.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.8 | 5.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.7 | 2.2 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.7 | 0.7 | GO:1903909 | regulation of receptor clustering(GO:1903909) regulation of neuromuscular junction development(GO:1904396) |
| 0.7 | 3.6 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.7 | 2.1 | GO:0021594 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.7 | 5.6 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.7 | 6.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.7 | 3.4 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.7 | 3.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.7 | 6.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.7 | 11.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 0.6 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.6 | 6.9 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.6 | 9.9 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.6 | 6.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.6 | 2.4 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.6 | 3.6 | GO:1901582 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.6 | 8.4 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.6 | 2.4 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.6 | 4.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.6 | 1.7 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.6 | 0.6 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.6 | 7.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.6 | 3.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.6 | 5.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.5 | 2.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.5 | 2.1 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.5 | 2.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.5 | 2.5 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.5 | 3.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.5 | 1.5 | GO:0006233 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.5 | 2.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.5 | 3.9 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.5 | 1.9 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.5 | 5.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.5 | 1.9 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.5 | 1.8 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.5 | 2.7 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.5 | 2.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.5 | 3.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.4 | 12.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.4 | 2.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.4 | 10.8 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.4 | 2.5 | GO:0014028 | notochord formation(GO:0014028) |
| 0.4 | 2.0 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.4 | 5.8 | GO:0030432 | peristalsis(GO:0030432) |
| 0.4 | 1.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.4 | 10.8 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.4 | 3.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 1.1 | GO:1903406 | regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.4 | 7.4 | GO:0007614 | short-term memory(GO:0007614) |
| 0.4 | 11.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 7.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.4 | 1.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.4 | 3.6 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 6.8 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.4 | 6.7 | GO:0001964 | startle response(GO:0001964) |
| 0.3 | 4.5 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.3 | 1.7 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.3 | 35.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 6.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 4.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 7.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.3 | 4.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.3 | 7.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.3 | 11.0 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 4.0 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 2.5 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.3 | 0.6 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.3 | 20.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.3 | 4.1 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.3 | 2.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 | 6.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.3 | 3.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 1.4 | GO:2001274 | positive regulation of fat cell proliferation(GO:0070346) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.3 | 2.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.3 | 1.3 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 3.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.3 | 5.9 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.3 | 8.5 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.3 | 2.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.3 | 6.1 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.3 | 2.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 3.6 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.3 | 1.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.3 | 6.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 0.7 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.2 | 6.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 5.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 2.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 1.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 1.0 | GO:0051938 | L-glutamate import(GO:0051938) regulation of intracellular mRNA localization(GO:1904580) |
| 0.2 | 0.2 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.2 | 0.7 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.2 | 7.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 5.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 2.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 2.7 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.2 | 1.3 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.2 | 6.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 4.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.2 | 1.0 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.2 | 0.8 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 1.4 | GO:0072710 | response to hydroxyurea(GO:0072710) |
| 0.2 | 1.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.2 | 2.8 | GO:0048512 | circadian behavior(GO:0048512) |
| 0.2 | 4.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 1.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 3.3 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 5.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 8.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 3.9 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.2 | 12.3 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.2 | 1.9 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 0.4 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.6 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.2 | 1.8 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.2 | 11.1 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.2 | 1.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 3.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 9.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.2 | 2.6 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.2 | 1.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 0.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.2 | 2.6 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 1.8 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 0.3 | GO:0043400 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) positive regulation of cortisol secretion(GO:0051464) |
| 0.2 | 15.4 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.2 | 9.5 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.1 | 0.7 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 6.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 2.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 2.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 6.2 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.1 | 1.2 | GO:1900042 | regulation of interleukin-2 secretion(GO:1900040) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 14.9 | GO:0015992 | proton transport(GO:0015992) |
| 0.1 | 1.0 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.8 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 12.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 4.0 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 1.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.6 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.1 | 6.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.1 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.1 | 1.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 1.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 5.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 2.7 | GO:0009268 | response to pH(GO:0009268) |
| 0.1 | 0.3 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 13.4 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.7 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 1.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.5 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 7.2 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 1.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 4.0 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.1 | 2.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 9.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 5.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 1.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 2.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.6 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 2.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 1.0 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 5.2 | GO:0043473 | pigmentation(GO:0043473) |
| 0.1 | 2.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 2.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 3.0 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 0.3 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 1.6 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 4.5 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.1 | 8.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 1.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.4 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 | 3.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.3 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 8.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.1 | 2.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.9 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 5.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 5.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.9 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.8 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.7 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 1.7 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 1.5 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 1.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.8 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 4.2 | GO:0050804 | modulation of synaptic transmission(GO:0050804) |
| 0.0 | 0.8 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 2.0 | GO:0055006 | cardiac cell development(GO:0055006) cardiac muscle cell development(GO:0055013) |
| 0.0 | 3.7 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 1.1 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 3.0 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.5 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.4 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 1.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.9 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.6 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.4 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.3 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.0 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.3 | GO:0060187 | cell pole(GO:0060187) |
| 3.4 | 13.7 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 3.0 | 15.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 3.0 | 26.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 2.5 | 7.4 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 2.1 | 10.4 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.9 | 11.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.7 | 24.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.3 | 4.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.2 | 7.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 1.1 | 15.9 | GO:0005883 | neurofilament(GO:0005883) |
| 1.1 | 3.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 1.1 | 5.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.1 | 1.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 1.1 | 10.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 1.0 | 3.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) PCSK9-AnxA2 complex(GO:1990667) |
| 1.0 | 8.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.9 | 24.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.9 | 25.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.8 | 42.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.8 | 10.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.8 | 18.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.8 | 4.8 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.8 | 11.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.8 | 4.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.7 | 14.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.7 | 3.9 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.6 | 6.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 3.6 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.6 | 20.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.6 | 2.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.5 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.5 | 12.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.5 | 63.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.5 | 2.4 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.5 | 3.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 17.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.5 | 3.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.5 | 5.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.5 | 1.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.4 | 4.0 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.4 | 6.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.4 | 16.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 1.7 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.4 | 3.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.4 | 7.1 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.4 | 1.5 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.4 | 3.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 3.6 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.4 | 6.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.3 | 4.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 8.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 8.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 1.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.3 | 2.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.3 | 4.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 8.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 6.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 3.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 2.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 108.2 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.3 | 0.8 | GO:0061474 | phagolysosome(GO:0032010) phagolysosome membrane(GO:0061474) |
| 0.3 | 20.0 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 0.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 7.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 6.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.7 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 3.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 1.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 14.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 1.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 1.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 0.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.2 | 14.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.2 | 0.7 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.2 | 2.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 6.2 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 25.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 4.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 4.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 5.9 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 7.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 16.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 4.2 | GO:0030904 | retromer complex(GO:0030904) early endosome membrane(GO:0031901) |
| 0.1 | 5.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 8.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 7.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 5.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 4.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 8.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 3.4 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.1 | 0.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 2.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 43.4 | GO:0044297 | cell body(GO:0044297) |
| 0.1 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 4.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.7 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 10.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 10.6 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 6.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 2.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 5.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 149.6 | GO:0031224 | intrinsic component of membrane(GO:0031224) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.7 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.3 | 43.0 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 6.7 | 20.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 5.5 | 16.5 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 5.4 | 26.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 5.1 | 15.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 4.7 | 14.0 | GO:0051424 | corticotropin-releasing hormone binding(GO:0051424) |
| 4.1 | 12.2 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 3.6 | 10.8 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 2.9 | 17.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 2.7 | 5.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.6 | 10.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.6 | 10.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 2.3 | 11.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 2.2 | 10.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.9 | 13.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 1.9 | 7.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.9 | 15.2 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.9 | 9.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.9 | 5.6 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.9 | 9.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 1.7 | 10.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 1.7 | 13.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.7 | 6.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 1.7 | 8.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 1.6 | 6.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 1.5 | 9.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.5 | 8.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 1.4 | 13.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.3 | 5.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 1.3 | 3.8 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 1.2 | 8.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.2 | 11.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 1.2 | 11.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.1 | 15.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 1.1 | 5.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.1 | 4.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 1.1 | 5.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.1 | 3.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 1.0 | 7.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.0 | 3.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 1.0 | 5.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.0 | 7.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 1.0 | 2.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.9 | 3.8 | GO:0038100 | nodal binding(GO:0038100) |
| 0.9 | 64.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.9 | 3.6 | GO:2001070 | starch binding(GO:2001070) |
| 0.9 | 3.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.9 | 13.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.9 | 7.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.9 | 26.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.8 | 6.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.8 | 5.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.8 | 12.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.8 | 2.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.8 | 15.7 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.8 | 7.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.8 | 2.4 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.8 | 7.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.8 | 20.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.7 | 3.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.7 | 2.9 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.7 | 2.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 4.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.7 | 6.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.7 | 8.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.7 | 3.9 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.6 | 8.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.6 | 6.3 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.6 | 2.5 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.6 | 2.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.6 | 4.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.6 | 5.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.6 | 9.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.6 | 2.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.6 | 8.5 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.6 | 3.9 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.6 | 3.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.6 | 7.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.6 | 3.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.6 | 11.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.5 | 4.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 22.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.5 | 6.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.5 | 3.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.5 | 6.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.5 | 1.5 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.5 | 2.0 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.5 | 2.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.5 | 11.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 16.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.5 | 1.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.4 | 5.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.4 | 19.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.4 | 1.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.4 | 5.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.4 | 8.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 2.4 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.4 | 17.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.4 | 7.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 2.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 8.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.4 | 3.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 5.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 5.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.4 | 2.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.4 | 11.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 14.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 6.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 16.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.3 | 1.0 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 24.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.3 | 8.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.3 | 15.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.3 | 6.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 5.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 6.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 3.8 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.3 | 1.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 4.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 1.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.3 | 9.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 8.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.3 | 14.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 0.7 | GO:0051538 | iron-responsive element binding(GO:0030350) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 1.7 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 1.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 9.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 3.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 5.8 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.2 | 12.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 0.7 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 1.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 12.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 3.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 10.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.2 | 6.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 1.7 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 2.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 1.5 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 0.6 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.2 | 6.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 13.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 0.7 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 1.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 6.3 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 3.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 3.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 2.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 15.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.4 | GO:0016401 | acetate CoA-transferase activity(GO:0008775) palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 1.9 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 2.8 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.8 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 3.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 6.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 4.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 2.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 3.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 18.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 21.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 7.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 2.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 1.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 15.1 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.6 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 2.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 11.5 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 3.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 4.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 11.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 5.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 42.7 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 7.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 6.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 4.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 2.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 4.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 2.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 6.3 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 9.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 1.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 4.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.5 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 26.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.2 | 3.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.8 | 20.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.6 | 29.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.6 | 14.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 25.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.5 | 10.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.4 | 21.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.3 | 17.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 12.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 11.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 10.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 7.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 5.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 2.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 6.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 6.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 47.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 9.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 5.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 4.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 7.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 27.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.2 | 29.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 1.1 | 25.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.1 | 28.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 1.0 | 15.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 1.0 | 17.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.0 | 15.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.9 | 16.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.8 | 19.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.8 | 7.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.7 | 7.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.7 | 23.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 18.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.5 | 10.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.5 | 33.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 16.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 7.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 11.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 3.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 4.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 5.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 13.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 8.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 4.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 5.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 5.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 7.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 4.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 3.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 3.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 3.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 8.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 37.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 5.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 3.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 7.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 1.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 5.8 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 7.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 6.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 6.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 4.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 11.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 4.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.7 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 6.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 3.1 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.5 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 1.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 14.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 4.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 3.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 3.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 9.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.0 | 2.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |