Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
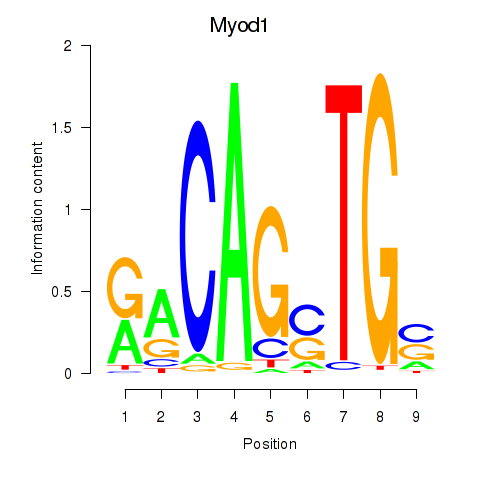
Results for Myod1
Z-value: 2.44
Transcription factors associated with Myod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myod1
|
ENSMUSG00000009471.5 | Myod1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myod1 | mm39_v1_chr7_+_46025890_46025904 | 0.85 | 5.8e-21 | Click! |
Activity profile of Myod1 motif
Sorted Z-values of Myod1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Myod1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_23629173 | 32.14 |
ENSMUST00000174435.2
|
Ank1
|
ankyrin 1, erythroid |
| chr8_+_23629046 | 30.87 |
ENSMUST00000121075.8
|
Ank1
|
ankyrin 1, erythroid |
| chr8_+_23629080 | 30.65 |
ENSMUST00000033947.15
|
Ank1
|
ankyrin 1, erythroid |
| chr7_+_126811831 | 27.25 |
ENSMUST00000127710.3
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr4_-_43523388 | 26.67 |
ENSMUST00000107913.10
ENSMUST00000030184.12 |
Tpm2
|
tropomyosin 2, beta |
| chr4_-_43523745 | 26.00 |
ENSMUST00000150592.2
|
Tpm2
|
tropomyosin 2, beta |
| chr2_-_52225763 | 25.99 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr2_+_152753231 | 24.22 |
ENSMUST00000028970.8
|
Mylk2
|
myosin, light polypeptide kinase 2, skeletal muscle |
| chr7_+_141995545 | 23.90 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr4_-_43523595 | 22.26 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr7_+_126810780 | 22.24 |
ENSMUST00000032910.13
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr7_+_19144950 | 22.22 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr2_-_113883285 | 21.66 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr6_-_97229708 | 20.37 |
ENSMUST00000095655.4
|
Lmod3
|
leiomodin 3 (fetal) |
| chr8_+_55003359 | 20.35 |
ENSMUST00000033918.4
|
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr7_-_140480314 | 20.33 |
ENSMUST00000026561.10
|
Cox8b
|
cytochrome c oxidase subunit 8B |
| chr1_+_75336965 | 18.15 |
ENSMUST00000027409.10
|
Des
|
desmin |
| chr18_-_60724855 | 17.86 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr7_+_45289391 | 17.78 |
ENSMUST00000148532.4
|
Mamstr
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr11_+_82802079 | 17.17 |
ENSMUST00000018989.14
ENSMUST00000164945.3 |
Unc45b
|
unc-45 myosin chaperone B |
| chr7_-_126046814 | 17.12 |
ENSMUST00000146973.2
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr17_-_48739874 | 16.65 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr7_+_141996067 | 15.66 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr3_-_116601451 | 15.15 |
ENSMUST00000159670.3
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
| chr7_-_44174065 | 15.01 |
ENSMUST00000165208.4
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr11_-_120441952 | 13.91 |
ENSMUST00000026121.3
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr2_-_164621641 | 13.58 |
ENSMUST00000103095.5
|
Tnnc2
|
troponin C2, fast |
| chr15_-_76906832 | 13.03 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr9_+_121606750 | 12.97 |
ENSMUST00000098272.4
|
Klhl40
|
kelch-like 40 |
| chr3_-_116601700 | 12.78 |
ENSMUST00000159742.8
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
| chr2_-_52225146 | 12.68 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr3_-_116601815 | 12.64 |
ENSMUST00000040603.14
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
| chrX_+_100492684 | 12.62 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr13_+_46571910 | 12.36 |
ENSMUST00000037923.5
|
Rbm24
|
RNA binding motif protein 24 |
| chr11_-_107607343 | 12.14 |
ENSMUST00000021065.6
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr9_-_50663571 | 11.84 |
ENSMUST00000042790.5
|
Hspb2
|
heat shock protein 2 |
| chrX_+_21581135 | 11.62 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr3_+_102981326 | 11.55 |
ENSMUST00000090715.13
ENSMUST00000155034.6 |
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr9_-_121621544 | 11.32 |
ENSMUST00000035110.11
|
Hhatl
|
hedgehog acyltransferase-like |
| chr10_-_80649315 | 11.31 |
ENSMUST00000181039.8
ENSMUST00000180438.2 |
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr1_+_135980639 | 11.24 |
ENSMUST00000112064.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr1_+_135980488 | 11.23 |
ENSMUST00000160641.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr17_-_27386763 | 11.22 |
ENSMUST00000025046.4
|
Ip6k3
|
inositol hexaphosphate kinase 3 |
| chr1_+_135980508 | 11.17 |
ENSMUST00000112068.10
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr4_+_129181407 | 11.11 |
ENSMUST00000102599.4
|
Sync
|
syncoilin |
| chr11_-_5753693 | 11.11 |
ENSMUST00000020768.4
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr9_+_50664288 | 11.06 |
ENSMUST00000214962.2
ENSMUST00000216755.2 |
Cryab
|
crystallin, alpha B |
| chr16_+_87350202 | 11.06 |
ENSMUST00000026700.8
|
Map3k7cl
|
Map3k7 C-terminal like |
| chr17_-_67939702 | 11.05 |
ENSMUST00000097290.4
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr14_+_55813074 | 10.81 |
ENSMUST00000022826.7
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr15_-_75620060 | 10.79 |
ENSMUST00000062002.6
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chr1_-_74788013 | 10.79 |
ENSMUST00000188073.7
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr8_+_46338557 | 10.60 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr5_-_69699965 | 10.37 |
ENSMUST00000031045.10
|
Yipf7
|
Yip1 domain family, member 7 |
| chr11_-_3477916 | 10.14 |
ENSMUST00000020718.10
|
Smtn
|
smoothelin |
| chr9_-_50663648 | 10.07 |
ENSMUST00000217159.2
|
Hspb2
|
heat shock protein 2 |
| chr3_+_102981352 | 10.01 |
ENSMUST00000176440.2
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr2_+_90865958 | 9.96 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr6_+_17307272 | 9.69 |
ENSMUST00000115454.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr9_-_119852624 | 9.52 |
ENSMUST00000111635.4
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr8_+_46338498 | 9.44 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr9_+_50664207 | 9.31 |
ENSMUST00000034562.9
|
Cryab
|
crystallin, alpha B |
| chr14_-_21798678 | 9.29 |
ENSMUST00000075040.9
|
Dusp13
|
dual specificity phosphatase 13 |
| chr2_-_84652890 | 9.27 |
ENSMUST00000028471.6
|
Smtnl1
|
smoothelin-like 1 |
| chr3_-_151971391 | 9.13 |
ENSMUST00000199470.5
ENSMUST00000200589.5 |
Nexn
|
nexilin |
| chr11_+_117700479 | 9.04 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr2_-_76812799 | 9.02 |
ENSMUST00000011934.13
ENSMUST00000099981.10 ENSMUST00000099980.10 ENSMUST00000111882.9 ENSMUST00000140091.8 |
Ttn
|
titin |
| chr5_-_69699932 | 8.80 |
ENSMUST00000202423.2
|
Yipf7
|
Yip1 domain family, member 7 |
| chr11_+_69856222 | 8.65 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chr10_-_81037878 | 8.58 |
ENSMUST00000005069.8
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr6_-_71239216 | 8.51 |
ENSMUST00000129630.3
ENSMUST00000114186.9 ENSMUST00000074301.10 |
Smyd1
|
SET and MYND domain containing 1 |
| chr7_+_28533279 | 8.32 |
ENSMUST00000208971.2
ENSMUST00000066723.15 |
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr6_+_17307639 | 8.22 |
ENSMUST00000115453.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr10_+_116822219 | 8.18 |
ENSMUST00000020378.5
|
Best3
|
bestrophin 3 |
| chr2_-_120370333 | 8.16 |
ENSMUST00000171215.8
|
Zfp106
|
zinc finger protein 106 |
| chr2_-_73410632 | 7.97 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr15_-_58078274 | 7.76 |
ENSMUST00000022986.8
|
Fbxo32
|
F-box protein 32 |
| chr11_-_59029996 | 7.73 |
ENSMUST00000219084.3
|
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr11_-_120538928 | 7.52 |
ENSMUST00000239158.2
ENSMUST00000026134.3 |
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr2_-_125348305 | 7.51 |
ENSMUST00000028633.13
|
Fbn1
|
fibrillin 1 |
| chr10_+_127337541 | 7.46 |
ENSMUST00000160019.8
ENSMUST00000160610.2 ENSMUST00000035839.3 |
Stac3
|
SH3 and cysteine rich domain 3 |
| chr7_+_27770655 | 7.22 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr8_-_106198112 | 7.20 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr11_+_48728291 | 7.17 |
ENSMUST00000046903.6
|
Trim7
|
tripartite motif-containing 7 |
| chr14_+_30601157 | 7.11 |
ENSMUST00000040715.8
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr14_-_21798694 | 6.95 |
ENSMUST00000183943.2
|
Dusp13
|
dual specificity phosphatase 13 |
| chr7_+_43874854 | 6.94 |
ENSMUST00000206144.2
|
Klk1
|
kallikrein 1 |
| chr10_+_3316505 | 6.90 |
ENSMUST00000217573.2
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr14_-_20714634 | 6.86 |
ENSMUST00000119483.2
|
Synpo2l
|
synaptopodin 2-like |
| chr11_+_110956980 | 6.72 |
ENSMUST00000042970.3
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr19_+_47167554 | 6.72 |
ENSMUST00000235290.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr9_+_43978290 | 6.70 |
ENSMUST00000034508.14
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr5_-_134776101 | 6.66 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr15_-_96953823 | 6.45 |
ENSMUST00000023101.10
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr11_+_96820220 | 6.28 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr8_-_120362291 | 6.26 |
ENSMUST00000061828.10
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr3_-_19319123 | 6.23 |
ENSMUST00000121951.2
|
Pde7a
|
phosphodiesterase 7A |
| chr19_+_5118103 | 6.21 |
ENSMUST00000070630.8
|
Cd248
|
CD248 antigen, endosialin |
| chr6_-_88604404 | 6.06 |
ENSMUST00000120933.5
|
Kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr19_+_7034149 | 6.05 |
ENSMUST00000040261.7
|
Macrod1
|
mono-ADP ribosylhydrolase 1 |
| chr3_+_60408600 | 5.96 |
ENSMUST00000099087.8
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr1_-_44258112 | 5.93 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E |
| chr8_-_120362085 | 5.92 |
ENSMUST00000164382.2
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chrX_-_73325318 | 5.85 |
ENSMUST00000239458.2
ENSMUST00000019232.10 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr2_+_153334710 | 5.83 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr16_+_57173632 | 5.64 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr4_-_104967032 | 5.64 |
ENSMUST00000030243.8
|
Prkaa2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr15_+_78810919 | 5.64 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr11_+_96820091 | 5.56 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr18_+_36481792 | 5.56 |
ENSMUST00000152804.9
|
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr10_+_36383008 | 5.55 |
ENSMUST00000168572.8
|
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr11_+_77353218 | 5.46 |
ENSMUST00000102493.8
|
Coro6
|
coronin 6 |
| chr18_+_36481706 | 5.43 |
ENSMUST00000235864.2
ENSMUST00000050584.10 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr9_+_43978369 | 5.28 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chrX_-_73325204 | 5.23 |
ENSMUST00000114189.10
ENSMUST00000119361.4 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr4_-_141351110 | 5.15 |
ENSMUST00000038661.8
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr10_-_23226034 | 5.14 |
ENSMUST00000219315.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr19_+_47167259 | 5.03 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr4_+_135487016 | 5.02 |
ENSMUST00000105854.2
|
Myom3
|
myomesin family, member 3 |
| chr11_+_87989972 | 5.00 |
ENSMUST00000018522.13
|
Cuedc1
|
CUE domain containing 1 |
| chr12_+_33004178 | 4.92 |
ENSMUST00000020885.13
|
Sypl
|
synaptophysin-like protein |
| chr1_+_40720731 | 4.87 |
ENSMUST00000192345.2
|
Slc9a2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
| chr11_+_106256298 | 4.84 |
ENSMUST00000009354.10
|
Prr29
|
proline rich 29 |
| chr3_-_144425819 | 4.84 |
ENSMUST00000199531.5
ENSMUST00000199854.5 |
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr11_-_70510010 | 4.83 |
ENSMUST00000102556.10
ENSMUST00000014753.9 |
Chrne
|
cholinergic receptor, nicotinic, epsilon polypeptide |
| chr13_-_49301407 | 4.79 |
ENSMUST00000162581.8
ENSMUST00000110097.9 ENSMUST00000049265.15 ENSMUST00000035538.13 ENSMUST00000110096.8 ENSMUST00000091623.10 |
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr7_+_99659121 | 4.74 |
ENSMUST00000107084.8
|
Chrdl2
|
chordin-like 2 |
| chr7_+_130294262 | 4.61 |
ENSMUST00000033141.7
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_+_17307038 | 4.61 |
ENSMUST00000123439.8
|
Cav1
|
caveolin 1, caveolae protein |
| chr17_+_48047955 | 4.54 |
ENSMUST00000086932.10
|
Tfeb
|
transcription factor EB |
| chr7_-_27010068 | 4.49 |
ENSMUST00000125455.2
|
Ltbp4
|
latent transforming growth factor beta binding protein 4 |
| chr13_-_60325170 | 4.49 |
ENSMUST00000065086.6
|
Gas1
|
growth arrest specific 1 |
| chr9_-_58066484 | 4.39 |
ENSMUST00000041477.15
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr3_-_63391300 | 4.36 |
ENSMUST00000192926.2
|
Strit1
|
small transmembrane regulator of ion transport 1 |
| chr1_+_165288606 | 4.36 |
ENSMUST00000027853.6
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr16_+_57173456 | 4.35 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr6_-_52168675 | 4.33 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4 |
| chr9_-_79884920 | 4.32 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1 |
| chr7_+_130294403 | 4.31 |
ENSMUST00000207282.2
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr2_+_122479770 | 4.31 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr11_-_54968901 | 4.30 |
ENSMUST00000055040.13
|
Ccdc69
|
coiled-coil domain containing 69 |
| chr8_-_123187406 | 4.30 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr6_-_124817155 | 4.22 |
ENSMUST00000024206.6
|
Gnb3
|
guanine nucleotide binding protein (G protein), beta 3 |
| chr9_-_79885063 | 4.20 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chr11_-_69496655 | 4.18 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr7_+_101916992 | 4.17 |
ENSMUST00000033289.6
ENSMUST00000209255.2 |
Stim1
|
stromal interaction molecule 1 |
| chr8_+_120121612 | 4.16 |
ENSMUST00000098367.5
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr3_+_60408678 | 4.14 |
ENSMUST00000191747.6
ENSMUST00000194069.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr1_-_75195889 | 4.12 |
ENSMUST00000186213.7
|
Tuba4a
|
tubulin, alpha 4A |
| chr1_-_75482975 | 4.07 |
ENSMUST00000113567.10
ENSMUST00000113565.3 |
Obsl1
|
obscurin-like 1 |
| chr11_-_101315345 | 4.04 |
ENSMUST00000107257.8
ENSMUST00000107259.4 ENSMUST00000107252.9 ENSMUST00000093933.11 |
Gm27029
Ptges3l
|
predicted gene, 27029 prostaglandin E synthase 3 like |
| chr6_-_99643723 | 4.04 |
ENSMUST00000032151.3
|
Eif4e3
|
eukaryotic translation initiation factor 4E member 3 |
| chr15_+_99615396 | 3.99 |
ENSMUST00000023760.13
ENSMUST00000162194.2 |
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr8_+_106245368 | 3.97 |
ENSMUST00000034363.7
|
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr7_+_142025575 | 3.97 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr7_+_3339059 | 3.94 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr1_+_87118346 | 3.91 |
ENSMUST00000073252.9
|
Chrnd
|
cholinergic receptor, nicotinic, delta polypeptide |
| chr18_+_32055339 | 3.88 |
ENSMUST00000233994.2
|
Lims2
|
LIM and senescent cell antigen like domains 2 |
| chr7_+_3339077 | 3.87 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr9_+_30853837 | 3.81 |
ENSMUST00000068135.13
|
Adamts8
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 8 |
| chr8_-_62355690 | 3.77 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr12_+_113104085 | 3.77 |
ENSMUST00000200380.5
|
Crip2
|
cysteine rich protein 2 |
| chr18_+_32510270 | 3.74 |
ENSMUST00000234857.2
ENSMUST00000234496.2 ENSMUST00000091967.13 ENSMUST00000025239.9 |
Bin1
|
bridging integrator 1 |
| chr12_+_113103817 | 3.72 |
ENSMUST00000084882.9
|
Crip2
|
cysteine rich protein 2 |
| chr14_+_67470884 | 3.62 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr6_+_112250719 | 3.62 |
ENSMUST00000032376.6
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr19_+_47167444 | 3.56 |
ENSMUST00000235326.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr2_-_84605732 | 3.53 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr2_-_131021905 | 3.52 |
ENSMUST00000089510.5
|
Cenpb
|
centromere protein B |
| chr4_-_107889136 | 3.52 |
ENSMUST00000106708.8
|
Podn
|
podocan |
| chrX_-_51702790 | 3.52 |
ENSMUST00000069360.14
|
Gpc3
|
glypican 3 |
| chr11_-_96714813 | 3.50 |
ENSMUST00000142065.2
ENSMUST00000167110.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr18_+_61096597 | 3.50 |
ENSMUST00000115295.9
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr19_-_45548942 | 3.50 |
ENSMUST00000026239.7
|
Poll
|
polymerase (DNA directed), lambda |
| chr2_-_84605764 | 3.50 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr9_-_58065800 | 3.49 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr4_+_58285957 | 3.46 |
ENSMUST00000081919.12
ENSMUST00000177951.8 ENSMUST00000098059.10 ENSMUST00000179951.2 ENSMUST00000102893.10 ENSMUST00000084578.12 ENSMUST00000098057.10 |
Musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr10_-_120037464 | 3.44 |
ENSMUST00000020448.11
|
Irak3
|
interleukin-1 receptor-associated kinase 3 |
| chrX_+_139857640 | 3.43 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr11_-_20781009 | 3.40 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr7_+_142025817 | 3.40 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr13_+_83652352 | 3.39 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr1_+_135764092 | 3.39 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr12_-_86125793 | 3.36 |
ENSMUST00000003687.8
|
Tgfb3
|
transforming growth factor, beta 3 |
| chr1_-_135302971 | 3.33 |
ENSMUST00000041240.4
|
Shisa4
|
shisa family member 4 |
| chr8_-_112417633 | 3.32 |
ENSMUST00000034435.7
|
Ctrb1
|
chymotrypsinogen B1 |
| chr12_-_76756772 | 3.30 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chrX_-_51702813 | 3.29 |
ENSMUST00000114857.2
|
Gpc3
|
glypican 3 |
| chr2_-_26962187 | 3.27 |
ENSMUST00000009358.9
|
Mymk
|
myomaker, myoblast fusion factor |
| chr11_+_50492899 | 3.25 |
ENSMUST00000142118.3
ENSMUST00000040523.9 |
Adamts2
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 2 |
| chr16_+_6887689 | 3.21 |
ENSMUST00000229741.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr14_+_67470735 | 3.20 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr13_+_83652150 | 3.17 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr1_-_172460497 | 3.16 |
ENSMUST00000027826.7
|
Dusp23
|
dual specificity phosphatase 23 |
| chrX_+_139857688 | 3.12 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr8_+_96078886 | 3.11 |
ENSMUST00000034243.7
|
Mmp15
|
matrix metallopeptidase 15 |
| chr11_+_102727122 | 3.08 |
ENSMUST00000021302.15
ENSMUST00000107072.2 |
Higd1b
|
HIG1 domain family, member 1B |
| chr2_-_62313981 | 3.01 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr4_+_15265798 | 3.00 |
ENSMUST00000062684.9
|
Tmem64
|
transmembrane protein 64 |
| chr3_+_89344006 | 2.96 |
ENSMUST00000038942.10
ENSMUST00000130858.8 ENSMUST00000146630.8 ENSMUST00000145753.2 |
Pbxip1
|
pre B cell leukemia transcription factor interacting protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 41.3 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 6.7 | 6.7 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 4.6 | 51.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 3.8 | 11.3 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 3.8 | 22.5 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 3.7 | 33.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 2.6 | 7.8 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 2.5 | 7.5 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 2.3 | 7.0 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 2.2 | 21.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 2.1 | 8.4 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 2.1 | 96.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 1.9 | 21.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 1.9 | 5.6 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.9 | 11.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.8 | 19.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.8 | 91.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 1.7 | 19.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.5 | 38.3 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 1.5 | 4.4 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 1.4 | 13.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.4 | 13.0 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 1.4 | 4.2 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 1.4 | 6.8 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 1.4 | 10.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.3 | 4.0 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 1.3 | 7.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 1.3 | 20.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.2 | 3.6 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.2 | 5.8 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 1.1 | 4.5 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 1.1 | 3.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 1.1 | 3.4 | GO:0003032 | detection of oxygen(GO:0003032) |
| 1.1 | 3.3 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 1.1 | 4.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 1.0 | 4.0 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.0 | 2.9 | GO:1903210 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 1.0 | 13.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.9 | 6.5 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.9 | 3.7 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.9 | 9.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.9 | 1.8 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.9 | 7.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.9 | 6.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.9 | 12.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.8 | 4.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.8 | 3.3 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.8 | 57.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.8 | 4.7 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.8 | 2.3 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.8 | 3.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.7 | 1.5 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.7 | 9.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.7 | 5.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.7 | 12.4 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.7 | 5.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.7 | 2.0 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.6 | 2.6 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.6 | 4.9 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.6 | 1.8 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.6 | 1.8 | GO:0043465 | regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.6 | 2.9 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.6 | 3.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.6 | 6.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.6 | 3.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.6 | 12.0 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.6 | 1.7 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of tolerance induction to self antigen(GO:0002649) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) endocardial cushion fusion(GO:0003274) |
| 0.6 | 3.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.6 | 1.7 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.6 | 1.7 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.5 | 1.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.5 | 2.7 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.5 | 1.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.5 | 2.7 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.5 | 17.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.5 | 4.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.5 | 3.6 | GO:0003383 | apical constriction(GO:0003383) |
| 0.5 | 2.1 | GO:0046340 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.5 | 24.5 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.5 | 12.1 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.5 | 8.6 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.5 | 14.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.4 | 12.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.4 | 2.7 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.4 | 1.3 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.4 | 2.6 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.4 | 2.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.4 | 4.9 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 2.0 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.4 | 2.4 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) |
| 0.4 | 2.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.4 | 2.3 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.4 | 2.7 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.4 | 1.9 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.4 | 10.8 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.4 | 8.9 | GO:0022027 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.4 | 1.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.4 | 1.8 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 | 3.9 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 1.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 3.4 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.3 | 4.6 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.3 | 11.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.3 | 4.5 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.3 | 9.5 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.3 | 0.6 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.3 | 67.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.3 | 2.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 6.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.3 | 0.3 | GO:0061324 | canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.3 | 0.9 | GO:0002588 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.3 | 6.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.3 | 0.8 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.3 | 2.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 1.4 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 | 2.5 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.3 | 0.8 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.3 | 4.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.3 | 0.8 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.3 | 0.8 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.3 | 10.8 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.3 | 10.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.3 | 3.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 12.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 1.5 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 1.7 | GO:0051541 | elastin metabolic process(GO:0051541) elastin catabolic process(GO:0060309) |
| 0.2 | 1.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 1.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 1.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 | 0.7 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.2 | 3.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 0.9 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.2 | 1.5 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.2 | 1.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 0.6 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) positive regulation of hindgut contraction(GO:0060450) regulation of relaxation of smooth muscle(GO:1901080) positive regulation of relaxation of smooth muscle(GO:1901082) |
| 0.2 | 2.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 | 2.4 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.2 | 4.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 17.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.2 | 1.0 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.2 | 1.7 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 1.2 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 58.3 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.2 | 4.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 3.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 1.5 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.2 | 0.8 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.2 | 1.6 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 0.5 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.2 | 2.6 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.2 | 0.8 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.2 | 8.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.9 | GO:0042637 | catagen(GO:0042637) |
| 0.1 | 2.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 1.0 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.1 | 0.6 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.5 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 1.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.9 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.5 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 1.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.3 | GO:0019087 | negative regulation of fibroblast migration(GO:0010764) transformation of host cell by virus(GO:0019087) |
| 0.1 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 2.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 1.6 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 1.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 4.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 7.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.6 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 1.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 1.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.5 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.4 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.6 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 4.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.6 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 2.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.6 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.3 | GO:0060538 | skeletal muscle organ development(GO:0060538) |
| 0.1 | 1.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 5.7 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.1 | 1.7 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 1.6 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.3 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.1 | GO:1903413 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.5 | GO:1901142 | insulin metabolic process(GO:1901142) |
| 0.1 | 0.4 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.1 | 0.9 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 4.5 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 7.6 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.1 | 0.6 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 2.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.5 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 5.0 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 3.8 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 2.2 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 1.9 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 2.8 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 1.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 1.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 10.6 | GO:0006520 | cellular amino acid metabolic process(GO:0006520) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 2.5 | GO:0001657 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 3.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.6 | GO:0010543 | regulation of platelet activation(GO:0010543) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 3.9 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.7 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.7 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 8.2 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.9 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.8 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.0 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.4 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.4 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 1.3 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 21.9 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 5.1 | 76.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 3.6 | 18.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.6 | 183.2 | GO:0031430 | M band(GO:0031430) |
| 3.0 | 56.5 | GO:0005861 | troponin complex(GO:0005861) |
| 2.3 | 38.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 2.1 | 17.1 | GO:0031673 | H zone(GO:0031673) |
| 1.3 | 6.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.3 | 22.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.3 | 24.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.1 | 4.6 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 1.0 | 2.9 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.8 | 17.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.8 | 84.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.8 | 2.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.6 | 5.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.6 | 13.8 | GO:0031672 | A band(GO:0031672) |
| 0.6 | 7.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.5 | 3.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 2.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.5 | 6.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.5 | 83.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.5 | 16.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 7.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 35.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.5 | 3.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 10.0 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.4 | 4.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 2.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 2.7 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.3 | 8.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 11.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 1.7 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 3.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 0.9 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 9.1 | GO:0030017 | sarcomere(GO:0030017) |
| 0.2 | 6.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 0.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 2.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 3.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 7.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 2.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 11.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 1.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 4.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 5.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 2.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 4.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 8.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 2.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 7.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 1.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 13.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 5.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 5.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 4.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 6.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 3.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 1.6 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 12.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 8.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 4.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 4.4 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 2.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.5 | 40.6 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 9.4 | 93.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 4.0 | 39.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 3.8 | 15.3 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 3.8 | 22.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 3.7 | 11.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 3.6 | 17.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 3.5 | 24.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 3.2 | 22.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 2.9 | 8.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 2.7 | 185.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 2.3 | 11.6 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 2.2 | 21.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.9 | 11.2 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 1.9 | 16.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 1.6 | 4.9 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 1.6 | 33.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.5 | 9.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.4 | 13.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.4 | 5.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.3 | 4.0 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 1.3 | 4.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.3 | 42.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.3 | 17.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 1.2 | 31.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.0 | 8.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.9 | 8.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.9 | 3.5 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.9 | 7.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.9 | 6.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.9 | 17.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.8 | 24.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.8 | 5.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.8 | 0.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.8 | 2.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.8 | 10.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.7 | 2.7 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.7 | 3.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.7 | 4.0 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.6 | 4.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.6 | 1.8 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.6 | 13.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.6 | 10.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 3.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 3.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.5 | 4.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 7.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.5 | 1.8 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.4 | 10.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 1.7 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.4 | 3.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.4 | 4.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 9.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 2.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 1.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 11.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 21.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.3 | 9.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 0.9 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 2.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.3 | 4.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 17.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.3 | 2.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 7.5 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.3 | 2.7 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.3 | 12.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 1.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 7.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 12.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 9.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 5.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 2.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 2.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 5.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 6.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 1.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 15.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 1.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 0.7 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 1.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 3.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 9.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 2.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 3.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 2.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 7.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.9 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 3.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 14.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 11.1 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.1 | 0.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 2.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 2.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.3 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 2.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 3.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 2.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.5 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 1.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 8.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 4.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 2.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 4.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 8.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 3.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 7.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.3 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 4.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 14.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 9.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 12.0 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.1 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 5.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 6.2 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.0 | 1.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 14.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 6.5 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.3 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 1.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 10.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.0 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 6.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 23.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.5 | 19.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.3 | 7.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 7.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 15.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 52.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 8.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 7.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 5.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 2.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 7.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 15.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 18.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 9.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 2.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 3.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 5.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 2.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 1.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 5.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 2.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 3.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 3.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 5.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 3.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 10.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 2.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 201.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.9 | 39.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 1.7 | 86.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.2 | 17.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 1.0 | 13.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.9 | 27.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.9 | 21.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.9 | 21.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.5 | 17.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.5 | 11.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 31.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 17.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 7.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 12.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 6.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 11.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 6.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 7.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 2.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.0 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.1 | 6.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 6.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.6 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 5.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 5.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 5.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 4.8 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 1.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 5.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.7 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 2.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 5.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 4.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 11.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.4 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |