Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
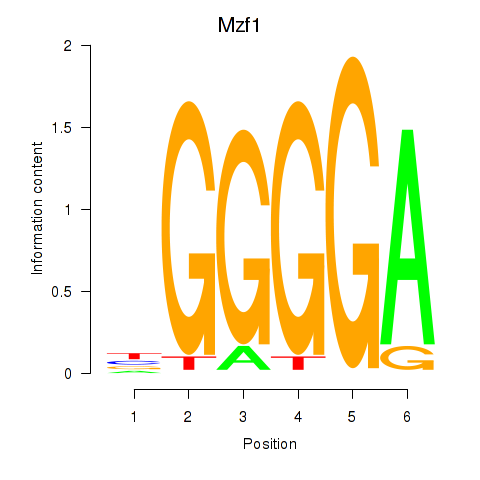
Results for Mzf1
Z-value: 1.74
Transcription factors associated with Mzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mzf1
|
ENSMUSG00000030380.18 | Mzf1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mzf1 | mm39_v1_chr7_-_12788441_12788441 | 0.66 | 2.0e-10 | Click! |
Activity profile of Mzf1 motif
Sorted Z-values of Mzf1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Mzf1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_69909245 | 19.46 |
ENSMUST00000231415.2
ENSMUST00000108588.9 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr3_-_117153802 | 14.15 |
ENSMUST00000197743.2
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr4_+_127066667 | 13.43 |
ENSMUST00000106094.9
|
Dlgap3
|
DLG associated protein 3 |
| chr18_+_34994253 | 12.20 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr11_+_69909659 | 10.72 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr11_+_69917640 | 10.17 |
ENSMUST00000135916.9
ENSMUST00000232659.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr2_+_121125918 | 9.83 |
ENSMUST00000110639.8
|
Map1a
|
microtubule-associated protein 1 A |
| chr1_-_123972900 | 9.75 |
ENSMUST00000112603.4
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr2_-_25471703 | 9.67 |
ENSMUST00000114217.3
ENSMUST00000191602.2 |
Ajm1
|
apical junction component 1 |
| chr6_+_110622533 | 9.19 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr7_+_45434755 | 9.18 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr12_-_72455708 | 9.08 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr11_-_42072990 | 8.92 |
ENSMUST00000205546.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr6_-_28831746 | 8.83 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr1_+_75483721 | 8.45 |
ENSMUST00000037330.5
|
Inha
|
inhibin alpha |
| chr8_+_106412905 | 7.67 |
ENSMUST00000213019.2
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr16_+_20513658 | 7.62 |
ENSMUST00000056518.13
|
Fam131a
|
family with sequence similarity 131, member A |
| chr4_+_127062924 | 7.53 |
ENSMUST00000046659.14
|
Dlgap3
|
DLG associated protein 3 |
| chr13_-_69887964 | 7.44 |
ENSMUST00000065118.7
|
Ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr3_-_86827664 | 7.39 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr15_+_101164719 | 7.36 |
ENSMUST00000230814.2
ENSMUST00000023779.8 |
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr5_-_115332343 | 7.31 |
ENSMUST00000112113.8
|
Cabp1
|
calcium binding protein 1 |
| chr11_+_94881861 | 7.08 |
ENSMUST00000038696.12
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr1_+_66360865 | 6.82 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr14_-_94128065 | 6.75 |
ENSMUST00000192221.6
ENSMUST00000195826.6 ENSMUST00000193901.6 ENSMUST00000194056.2 |
Pcdh9
|
protocadherin 9 |
| chr7_+_4693759 | 6.67 |
ENSMUST00000048248.9
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr14_+_55747902 | 6.63 |
ENSMUST00000165262.8
ENSMUST00000074225.11 |
Cpne6
|
copine VI |
| chr7_+_4693603 | 6.52 |
ENSMUST00000120836.8
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr11_-_97913420 | 6.47 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr3_-_88669551 | 6.46 |
ENSMUST00000183267.2
|
Syt11
|
synaptotagmin XI |
| chr2_-_19002932 | 6.28 |
ENSMUST00000006912.12
|
Pip4k2a
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr7_-_78228116 | 6.15 |
ENSMUST00000206268.2
ENSMUST00000039431.14 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr3_+_45332831 | 6.10 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr3_+_96088467 | 6.04 |
ENSMUST00000035371.9
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr19_+_38252984 | 5.98 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr11_-_42073737 | 5.86 |
ENSMUST00000206085.2
ENSMUST00000020707.12 ENSMUST00000132971.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr16_+_91066602 | 5.86 |
ENSMUST00000056882.7
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr16_-_34083549 | 5.85 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr1_-_132635042 | 5.83 |
ENSMUST00000043189.14
|
Nfasc
|
neurofascin |
| chr18_-_43032359 | 5.69 |
ENSMUST00000117687.8
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr14_+_10123804 | 5.64 |
ENSMUST00000022262.6
ENSMUST00000224714.2 |
Fezf2
|
Fez family zinc finger 2 |
| chr6_-_42301574 | 5.50 |
ENSMUST00000031891.15
ENSMUST00000143278.8 |
Fam131b
|
family with sequence similarity 131, member B |
| chr12_+_105302853 | 5.45 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr1_-_154602102 | 5.42 |
ENSMUST00000187541.7
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr14_+_66581818 | 5.42 |
ENSMUST00000118426.8
ENSMUST00000121955.8 ENSMUST00000120229.8 ENSMUST00000134440.2 |
Stmn4
|
stathmin-like 4 |
| chr5_-_38316706 | 5.39 |
ENSMUST00000201341.2
ENSMUST00000201363.4 ENSMUST00000201134.2 |
Nsg1
|
neuron specific gene family member 1 |
| chr1_-_132635078 | 5.36 |
ENSMUST00000187861.7
|
Nfasc
|
neurofascin |
| chrX_-_46981379 | 5.35 |
ENSMUST00000077569.11
ENSMUST00000101616.9 ENSMUST00000088973.11 |
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr17_-_91396154 | 5.34 |
ENSMUST00000161402.10
ENSMUST00000054059.15 ENSMUST00000072671.14 ENSMUST00000174331.8 |
Nrxn1
|
neurexin I |
| chr1_-_123973223 | 5.32 |
ENSMUST00000112606.8
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr16_-_34083315 | 5.29 |
ENSMUST00000114953.8
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr11_-_42072920 | 5.28 |
ENSMUST00000207274.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr2_+_163280375 | 5.25 |
ENSMUST00000109420.10
ENSMUST00000109421.10 ENSMUST00000018087.13 ENSMUST00000137070.2 |
Gdap1l1
|
ganglioside-induced differentiation-associated protein 1-like 1 |
| chr15_-_8740218 | 5.24 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr19_+_38253077 | 5.24 |
ENSMUST00000198045.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr11_-_69451012 | 5.23 |
ENSMUST00000004036.6
|
Efnb3
|
ephrin B3 |
| chr5_-_113957362 | 5.20 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr17_+_37361523 | 5.19 |
ENSMUST00000172792.8
ENSMUST00000174347.2 |
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr16_-_28571820 | 5.10 |
ENSMUST00000232352.2
|
Fgf12
|
fibroblast growth factor 12 |
| chr2_+_129435115 | 5.09 |
ENSMUST00000099113.10
ENSMUST00000103202.10 |
Sirpa
|
signal-regulatory protein alpha |
| chr10_-_116309764 | 5.09 |
ENSMUST00000068233.11
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr7_-_105230395 | 5.06 |
ENSMUST00000188726.2
ENSMUST00000188440.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr3_-_80710097 | 5.06 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr18_-_43032514 | 5.04 |
ENSMUST00000236238.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr14_+_66581745 | 4.99 |
ENSMUST00000152093.8
ENSMUST00000074523.13 |
Stmn4
|
stathmin-like 4 |
| chr7_-_24771717 | 4.97 |
ENSMUST00000003468.10
|
Grik5
|
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| chr15_-_8739893 | 4.95 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr19_+_48194464 | 4.91 |
ENSMUST00000078880.6
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr19_+_38253105 | 4.90 |
ENSMUST00000196090.2
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr6_-_114018982 | 4.89 |
ENSMUST00000101045.10
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr2_+_70392351 | 4.89 |
ENSMUST00000094934.11
|
Gad1
|
glutamate decarboxylase 1 |
| chr3_+_96503944 | 4.89 |
ENSMUST00000058943.8
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr5_-_38316296 | 4.89 |
ENSMUST00000201415.4
|
Nsg1
|
neuron specific gene family member 1 |
| chr11_+_16207705 | 4.83 |
ENSMUST00000109645.9
ENSMUST00000109647.3 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chrX_+_47608122 | 4.83 |
ENSMUST00000033430.3
|
Rab33a
|
RAB33A, member RAS oncogene family |
| chr18_-_43032535 | 4.79 |
ENSMUST00000120632.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr4_+_129878627 | 4.78 |
ENSMUST00000120204.8
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr11_+_69920542 | 4.78 |
ENSMUST00000232266.2
ENSMUST00000132597.5 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr10_-_42459624 | 4.78 |
ENSMUST00000019938.11
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr4_-_46991842 | 4.74 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr3_-_87934772 | 4.72 |
ENSMUST00000005014.9
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr18_-_25886750 | 4.69 |
ENSMUST00000224553.2
ENSMUST00000025117.14 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr7_+_101070897 | 4.67 |
ENSMUST00000163751.10
ENSMUST00000211368.2 ENSMUST00000166652.2 |
Pde2a
|
phosphodiesterase 2A, cGMP-stimulated |
| chr14_-_124914516 | 4.62 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chr3_+_94385602 | 4.62 |
ENSMUST00000199884.5
ENSMUST00000198316.5 ENSMUST00000197558.5 |
Celf3
|
CUGBP, Elav-like family member 3 |
| chr6_-_42301488 | 4.62 |
ENSMUST00000095974.4
|
Fam131b
|
family with sequence similarity 131, member B |
| chrX_+_40490005 | 4.60 |
ENSMUST00000115103.9
ENSMUST00000076349.12 |
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr6_+_115111860 | 4.59 |
ENSMUST00000169345.4
|
Syn2
|
synapsin II |
| chr5_-_31250817 | 4.56 |
ENSMUST00000031037.14
|
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr2_+_119629995 | 4.56 |
ENSMUST00000028763.10
|
Tyro3
|
TYRO3 protein tyrosine kinase 3 |
| chr19_+_7245591 | 4.56 |
ENSMUST00000066646.12
|
Rcor2
|
REST corepressor 2 |
| chr7_+_29991366 | 4.54 |
ENSMUST00000144508.2
|
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr6_-_85479840 | 4.50 |
ENSMUST00000161546.2
ENSMUST00000161078.8 |
Fbxo41
|
F-box protein 41 |
| chr16_-_9812787 | 4.49 |
ENSMUST00000199708.5
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr16_-_34083200 | 4.43 |
ENSMUST00000114947.2
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr5_+_130477642 | 4.41 |
ENSMUST00000111288.4
|
Caln1
|
calneuron 1 |
| chr14_-_39194782 | 4.40 |
ENSMUST00000168810.9
ENSMUST00000173780.2 ENSMUST00000166968.9 |
Nrg3
|
neuregulin 3 |
| chr11_-_97635484 | 4.39 |
ENSMUST00000018691.9
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr5_+_37025810 | 4.39 |
ENSMUST00000031003.11
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr17_+_47451868 | 4.37 |
ENSMUST00000190080.9
|
Trerf1
|
transcriptional regulating factor 1 |
| chr2_+_129434738 | 4.37 |
ENSMUST00000153491.8
ENSMUST00000161620.8 ENSMUST00000179001.8 |
Sirpa
|
signal-regulatory protein alpha |
| chr7_-_105230479 | 4.35 |
ENSMUST00000191601.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr16_-_45830575 | 4.35 |
ENSMUST00000130481.2
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr5_-_138262178 | 4.34 |
ENSMUST00000048421.14
|
Map11
|
microtubule associated protein 11 |
| chr11_+_69920956 | 4.30 |
ENSMUST00000232115.2
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr2_-_17735847 | 4.28 |
ENSMUST00000028080.12
|
Nebl
|
nebulette |
| chr3_-_86827640 | 4.28 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr11_-_100650768 | 4.27 |
ENSMUST00000107363.3
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr2_+_129434834 | 4.27 |
ENSMUST00000103203.8
|
Sirpa
|
signal-regulatory protein alpha |
| chr11_-_4897991 | 4.26 |
ENSMUST00000093369.5
|
Nefh
|
neurofilament, heavy polypeptide |
| chr16_-_20440005 | 4.25 |
ENSMUST00000052939.4
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr4_-_68872585 | 4.22 |
ENSMUST00000030036.6
|
Brinp1
|
bone morphogenic protein/retinoic acid inducible neural specific 1 |
| chr3_-_89152320 | 4.22 |
ENSMUST00000107464.8
ENSMUST00000090924.13 |
Trim46
|
tripartite motif-containing 46 |
| chr16_+_32480040 | 4.21 |
ENSMUST00000238806.2
ENSMUST00000238856.2 |
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr15_+_101122069 | 4.21 |
ENSMUST00000000543.6
|
Tamalin
|
trafficking regulator and scaffold protein tamalin |
| chr5_-_137739364 | 4.19 |
ENSMUST00000149512.3
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr9_+_89791943 | 4.18 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr11_+_103061905 | 4.18 |
ENSMUST00000042286.12
ENSMUST00000218163.2 |
Fmnl1
|
formin-like 1 |
| chr18_+_37952556 | 4.17 |
ENSMUST00000055935.11
|
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr12_+_81678002 | 4.16 |
ENSMUST00000218362.2
|
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr2_+_97298002 | 4.15 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr8_-_9821021 | 4.11 |
ENSMUST00000208933.2
ENSMUST00000110969.5 |
Fam155a
|
family with sequence similarity 155, member A |
| chr9_-_56542908 | 4.08 |
ENSMUST00000114256.2
|
Lingo1
|
leucine rich repeat and Ig domain containing 1 |
| chr10_-_127024641 | 4.06 |
ENSMUST00000218654.2
|
Arhgef25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr1_+_128031055 | 4.03 |
ENSMUST00000188381.7
ENSMUST00000187900.7 ENSMUST00000036288.11 |
R3hdm1
|
R3H domain containing 1 |
| chr2_+_31135813 | 4.02 |
ENSMUST00000000199.8
|
Ncs1
|
neuronal calcium sensor 1 |
| chr2_+_71884943 | 4.00 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr5_+_137059127 | 3.98 |
ENSMUST00000041543.9
ENSMUST00000186451.2 |
Vgf
|
VGF nerve growth factor inducible |
| chrX_-_94209913 | 3.97 |
ENSMUST00000113873.9
ENSMUST00000113876.9 ENSMUST00000199920.5 ENSMUST00000113885.8 ENSMUST00000113883.8 ENSMUST00000196012.2 ENSMUST00000182001.8 ENSMUST00000113878.8 ENSMUST00000113882.8 ENSMUST00000182562.2 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr11_-_97464866 | 3.96 |
ENSMUST00000207653.2
ENSMUST00000107593.8 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr14_+_55131568 | 3.95 |
ENSMUST00000116476.9
ENSMUST00000022808.14 ENSMUST00000150975.8 |
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr7_+_126376353 | 3.94 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr11_+_17001818 | 3.91 |
ENSMUST00000058159.6
|
Cnrip1
|
cannabinoid receptor interacting protein 1 |
| chr7_-_78227518 | 3.91 |
ENSMUST00000195262.6
ENSMUST00000193002.6 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr11_+_87651359 | 3.87 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr7_-_30750856 | 3.87 |
ENSMUST00000073892.6
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr11_-_98220466 | 3.84 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr13_-_117161921 | 3.82 |
ENSMUST00000223949.2
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chrX_-_46981273 | 3.82 |
ENSMUST00000153548.9
ENSMUST00000141084.3 |
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr2_+_174127145 | 3.82 |
ENSMUST00000130761.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr12_+_102915709 | 3.81 |
ENSMUST00000179002.8
|
Unc79
|
unc-79 homolog |
| chr4_+_129878890 | 3.80 |
ENSMUST00000106017.8
ENSMUST00000121049.8 |
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr5_-_108515740 | 3.80 |
ENSMUST00000197216.3
|
Gm42517
|
predicted gene 42517 |
| chr18_-_61147272 | 3.78 |
ENSMUST00000025520.10
|
Slc6a7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 |
| chr3_+_102642516 | 3.78 |
ENSMUST00000119902.6
|
Tspan2
|
tetraspanin 2 |
| chr7_+_91321694 | 3.74 |
ENSMUST00000238608.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr7_-_30750828 | 3.74 |
ENSMUST00000206341.2
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr19_+_4771089 | 3.71 |
ENSMUST00000238976.3
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr18_+_34973605 | 3.69 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr4_+_102617495 | 3.68 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr13_+_42862957 | 3.67 |
ENSMUST00000066928.12
ENSMUST00000148891.8 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr7_+_27222678 | 3.66 |
ENSMUST00000108353.9
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr7_-_127410096 | 3.66 |
ENSMUST00000156135.3
|
Stx1b
|
syntaxin 1B |
| chr10_+_112292161 | 3.65 |
ENSMUST00000219607.2
ENSMUST00000218827.2 |
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr7_-_105217851 | 3.63 |
ENSMUST00000188368.7
ENSMUST00000187057.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chrX_+_133195974 | 3.63 |
ENSMUST00000037687.8
|
Tmem35a
|
transmembrane protein 35A |
| chr2_-_165076609 | 3.62 |
ENSMUST00000065438.13
|
Cdh22
|
cadherin 22 |
| chr15_+_99590098 | 3.61 |
ENSMUST00000228185.2
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr2_-_13016570 | 3.58 |
ENSMUST00000061545.7
|
C1ql3
|
C1q-like 3 |
| chr16_+_11802445 | 3.57 |
ENSMUST00000170672.9
ENSMUST00000023138.8 |
Shisa9
|
shisa family member 9 |
| chr15_+_98532624 | 3.57 |
ENSMUST00000003442.9
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr7_+_44078366 | 3.51 |
ENSMUST00000127790.8
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr11_+_115044966 | 3.51 |
ENSMUST00000021076.6
|
Rab37
|
RAB37, member RAS oncogene family |
| chr11_-_97520511 | 3.50 |
ENSMUST00000052281.6
|
Epop
|
elongin BC and polycomb repressive complex 2 associated protein |
| chr9_+_45341589 | 3.49 |
ENSMUST00000239471.2
ENSMUST00000034592.11 ENSMUST00000239429.2 |
Dscaml1
|
DS cell adhesion molecule like 1 |
| chr11_+_24030663 | 3.48 |
ENSMUST00000118955.2
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr3_+_107008343 | 3.47 |
ENSMUST00000197470.5
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr7_+_126376099 | 3.44 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr4_-_43578823 | 3.43 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2 |
| chr17_+_9068805 | 3.43 |
ENSMUST00000115720.8
|
Pde10a
|
phosphodiesterase 10A |
| chr15_+_88943916 | 3.43 |
ENSMUST00000161372.2
ENSMUST00000162424.2 |
Panx2
|
pannexin 2 |
| chr19_-_45804446 | 3.42 |
ENSMUST00000079431.10
ENSMUST00000026247.13 ENSMUST00000162528.9 |
Kcnip2
|
Kv channel-interacting protein 2 |
| chr7_-_57036920 | 3.42 |
ENSMUST00000068911.13
|
Gabrg3
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 3 |
| chr6_-_138399896 | 3.42 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chrX_-_58211440 | 3.41 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr9_-_70048766 | 3.41 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr1_+_140173787 | 3.40 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr12_+_88689638 | 3.39 |
ENSMUST00000190626.7
ENSMUST00000167103.8 |
Nrxn3
|
neurexin III |
| chr11_+_70323452 | 3.38 |
ENSMUST00000084954.13
ENSMUST00000108568.10 ENSMUST00000079056.9 ENSMUST00000102564.11 ENSMUST00000124943.8 ENSMUST00000150076.8 ENSMUST00000102563.2 |
Arrb2
|
arrestin, beta 2 |
| chr8_-_124586159 | 3.38 |
ENSMUST00000034452.12
|
Ccsap
|
centriole, cilia and spindle associated protein |
| chr4_+_155976279 | 3.38 |
ENSMUST00000105584.10
ENSMUST00000079031.6 |
Acap3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr15_-_25413838 | 3.37 |
ENSMUST00000058845.9
|
Basp1
|
brain abundant, membrane attached signal protein 1 |
| chr10_-_81308693 | 3.36 |
ENSMUST00000147524.3
ENSMUST00000119060.8 |
Celf5
|
CUGBP, Elav-like family member 5 |
| chr8_+_84627332 | 3.34 |
ENSMUST00000045393.15
ENSMUST00000132500.8 ENSMUST00000152978.8 |
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr3_+_28835425 | 3.34 |
ENSMUST00000060500.9
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr7_-_105230807 | 3.33 |
ENSMUST00000191011.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr6_-_13838423 | 3.31 |
ENSMUST00000115492.2
|
Gpr85
|
G protein-coupled receptor 85 |
| chr12_-_100486950 | 3.30 |
ENSMUST00000223020.2
ENSMUST00000062957.8 |
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr2_+_102489558 | 3.30 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr5_-_31453206 | 3.30 |
ENSMUST00000041266.11
ENSMUST00000172435.8 ENSMUST00000201417.2 |
Fndc4
|
fibronectin type III domain containing 4 |
| chrX_+_165127688 | 3.29 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr14_-_70864448 | 3.28 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr3_+_102642272 | 3.27 |
ENSMUST00000196611.5
|
Tspan2
|
tetraspanin 2 |
| chr15_+_99122742 | 3.26 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr11_+_69920849 | 3.25 |
ENSMUST00000143920.4
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr5_-_36555434 | 3.25 |
ENSMUST00000037370.14
ENSMUST00000070720.8 |
Sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr11_-_100650566 | 3.25 |
ENSMUST00000107361.9
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr5_-_8417982 | 3.24 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr4_+_102617332 | 3.23 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr13_-_117162041 | 3.23 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 3.6 | 21.8 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 3.6 | 50.9 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 2.6 | 15.6 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 2.6 | 7.7 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 2.4 | 7.1 | GO:2000474 | cellular response to morphine(GO:0071315) regulation of opioid receptor signaling pathway(GO:2000474) |
| 2.2 | 15.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.0 | 12.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 2.0 | 10.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 2.0 | 10.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 1.9 | 5.8 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 1.9 | 7.8 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 1.8 | 10.6 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 1.7 | 8.4 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 1.5 | 12.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.4 | 4.3 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 1.4 | 4.3 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 1.3 | 3.8 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 1.2 | 3.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 1.2 | 12.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.1 | 9.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 1.1 | 4.4 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 1.1 | 6.5 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.1 | 3.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 1.0 | 5.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.0 | 3.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 1.0 | 10.3 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 1.0 | 5.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.0 | 23.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.0 | 6.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.0 | 4.0 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 1.0 | 11.9 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 1.0 | 7.8 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 1.0 | 1.9 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.9 | 1.9 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.9 | 3.8 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.9 | 5.6 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.9 | 4.7 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.9 | 6.3 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.9 | 2.7 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.9 | 5.3 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.9 | 10.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.9 | 5.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.9 | 3.5 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.9 | 15.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.9 | 5.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.9 | 4.3 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.8 | 4.2 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.8 | 3.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.8 | 3.2 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.8 | 4.8 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.8 | 6.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.8 | 5.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.8 | 2.3 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.8 | 3.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.8 | 6.0 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.7 | 11.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.7 | 4.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.7 | 2.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.7 | 5.7 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.7 | 2.8 | GO:0021679 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) |
| 0.7 | 3.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.7 | 4.9 | GO:0048840 | otolith development(GO:0048840) |
| 0.7 | 2.7 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.7 | 1.3 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.7 | 19.7 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.6 | 3.8 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.6 | 8.8 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.6 | 1.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.6 | 3.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.6 | 3.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.6 | 3.7 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.6 | 1.8 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.6 | 5.4 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.6 | 6.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.6 | 2.4 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.6 | 4.7 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.6 | 2.3 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.6 | 4.5 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.6 | 5.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.6 | 5.0 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.6 | 7.2 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.5 | 10.9 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.5 | 2.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.5 | 1.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.5 | 2.1 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.5 | 38.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.5 | 2.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.5 | 4.6 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.5 | 3.9 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.5 | 8.3 | GO:0043084 | penile erection(GO:0043084) |
| 0.5 | 1.5 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.5 | 3.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.5 | 6.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.5 | 10.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.5 | 2.9 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.5 | 6.6 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.5 | 1.4 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.5 | 1.9 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.5 | 1.4 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.4 | 0.9 | GO:2000282 | regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.4 | 9.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 1.3 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.4 | 3.9 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.4 | 3.9 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.4 | 7.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.4 | 2.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 1.3 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 | 5.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.4 | 0.8 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.4 | 1.7 | GO:0072021 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) DCT cell differentiation(GO:0072069) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) metanephric DCT cell differentiation(GO:0072240) |
| 0.4 | 2.5 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.4 | 3.3 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 3.3 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.4 | 2.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 1.2 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.4 | 14.7 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.4 | 1.2 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.4 | 1.1 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.4 | 1.5 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.4 | 2.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.4 | 3.4 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.4 | 14.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.4 | 1.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 | 3.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 1.1 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.3 | 2.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 1.7 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.3 | 1.4 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 1.0 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.3 | 4.7 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.3 | 2.0 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.3 | 1.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.3 | 1.9 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.3 | 2.8 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 8.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.3 | 2.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 12.0 | GO:1900449 | regulation of neurotransmitter receptor activity(GO:0099601) regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.3 | 0.6 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.3 | 6.8 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.3 | 15.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.3 | 1.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.3 | 1.7 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.3 | 1.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.3 | 1.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.3 | 3.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 5.3 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.3 | 2.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 3.9 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.3 | 0.8 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.3 | 4.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 2.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 1.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.3 | 2.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.3 | 1.3 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.3 | 1.8 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.3 | 1.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.3 | 0.8 | GO:0070318 | enucleate erythrocyte development(GO:0048822) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.3 | 1.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.3 | 2.5 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.3 | 2.5 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.3 | 1.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 2.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.2 | 1.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 1.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 1.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 14.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 4.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 1.5 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.2 | 1.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 0.7 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.2 | 4.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 1.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 0.7 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.2 | 0.9 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 7.2 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.2 | 4.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 2.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 1.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 14.8 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.2 | 1.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 1.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 2.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 0.4 | GO:0071640 | macrophage inflammatory protein-1 alpha production(GO:0071608) regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.2 | 1.3 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.2 | 2.8 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.2 | 1.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 4.0 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 2.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 4.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.2 | 1.2 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 | 3.7 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.2 | 7.0 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.2 | 4.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.2 | 1.8 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 1.6 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.2 | 2.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.4 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.2 | 1.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 1.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 0.8 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 2.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 5.0 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.2 | 8.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 1.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 19.7 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.2 | 1.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.2 | 0.6 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.2 | 1.4 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 1.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 14.8 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.2 | 2.9 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.2 | 17.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 2.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 0.7 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 | 4.2 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.2 | 1.5 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.2 | 0.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 0.8 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.2 | 3.0 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.2 | 0.3 | GO:0060300 | regulation of cytokine activity(GO:0060300) positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 2.7 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 1.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 2.8 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 4.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 5.5 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 6.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.8 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 3.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.4 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 2.2 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.1 | 3.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 5.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 6.8 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 3.0 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.4 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 3.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 4.1 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.1 | 0.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 4.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.9 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 2.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 2.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.8 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.1 | 2.7 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 1.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 2.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.4 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 1.0 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 3.3 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 11.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 1.2 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 3.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 3.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 6.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 2.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 9.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.0 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.6 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 1.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.3 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.1 | 0.7 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.6 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 2.9 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 1.8 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 1.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.3 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.1 | 0.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 2.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 1.2 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.3 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.7 | GO:0060717 | chorion development(GO:0060717) |
| 0.1 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 1.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.1 | 1.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 1.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 1.7 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 1.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.7 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 1.4 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 2.2 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.1 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 2.0 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 2.2 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 1.2 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.2 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) |
| 0.0 | 0.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 1.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 1.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.7 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.6 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.0 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.0 | GO:0002442 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) platelet degranulation(GO:0002576) |
| 0.0 | 0.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.3 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 15.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 2.8 | 8.4 | GO:0043512 | inhibin A complex(GO:0043512) |
| 2.8 | 75.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 2.5 | 7.6 | GO:0090537 | CERF complex(GO:0090537) |
| 2.2 | 11.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 2.1 | 6.2 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 2.0 | 21.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 1.4 | 7.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 1.4 | 25.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.1 | 19.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.1 | 4.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.9 | 6.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.8 | 23.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.8 | 4.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.8 | 10.3 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.8 | 7.7 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.8 | 3.8 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.8 | 6.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.7 | 7.4 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.6 | 5.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.6 | 3.7 | GO:0070449 | elongin complex(GO:0070449) |
| 0.6 | 7.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.6 | 2.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.5 | 14.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 16.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 3.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.5 | 4.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.5 | 2.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.5 | 1.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.5 | 3.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 6.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 1.3 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.4 | 1.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.4 | 2.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 1.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.4 | 1.6 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.4 | 15.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 4.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.4 | 40.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.4 | 8.1 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.4 | 1.1 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.3 | 11.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 4.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 2.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 3.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 6.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.3 | 5.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 5.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 6.0 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 4.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 13.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 3.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 1.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.3 | 16.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 7.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 1.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 101.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.2 | 0.7 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.2 | 3.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 4.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 4.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.2 | 6.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 3.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 1.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 1.9 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 0.9 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 5.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 0.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 3.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 2.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 4.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 19.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.2 | 1.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 2.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 14.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 6.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 6.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 8.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.3 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 1.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 4.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 1.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 21.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 2.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 14.8 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 2.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 4.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.7 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 2.1 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 4.3 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 12.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 3.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 2.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 3.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 2.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 6.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 3.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 5.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 2.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 9.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 52.7 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 3.6 | 10.7 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 2.6 | 15.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.5 | 20.1 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 2.3 | 9.2 | GO:0070905 | serine binding(GO:0070905) |
| 2.2 | 13.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 2.1 | 12.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.1 | 6.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 1.7 | 12.2 | GO:0044729 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 1.6 | 4.7 | GO:0036004 | GAF domain binding(GO:0036004) |
| 1.4 | 4.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 1.1 | 13.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.1 | 3.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 1.1 | 12.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.0 | 3.1 | GO:0043404 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) corticotropin-releasing hormone binding(GO:0051424) |
| 1.0 | 5.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.0 | 9.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.0 | 10.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 1.0 | 6.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.0 | 5.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.0 | 3.9 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.9 | 14.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.9 | 17.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.9 | 5.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.9 | 4.5 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.9 | 4.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.9 | 7.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.9 | 7.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.9 | 10.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.8 | 9.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.8 | 4.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.8 | 4.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.8 | 5.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.8 | 9.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.7 | 2.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.7 | 2.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.7 | 4.9 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.7 | 4.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.7 | 3.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.7 | 17.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.6 | 1.9 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.6 | 12.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.6 | 1.9 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.6 | 19.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.6 | 4.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.6 | 2.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.6 | 2.3 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.5 | 3.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.5 | 3.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 2.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.5 | 4.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.5 | 5.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.5 | 2.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.5 | 1.5 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.5 | 1.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.5 | 17.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.5 | 2.3 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.4 | 1.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 1.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.4 | 3.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 5.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.4 | 1.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.4 | 4.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 2.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 1.2 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.4 | 6.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.4 | 3.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.4 | 1.1 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.4 | 1.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 1.8 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.4 | 6.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 1.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.3 | 1.0 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.3 | 2.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 6.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 2.7 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.3 | 13.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 19.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 18.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.3 | 1.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.3 | 1.8 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.3 | 2.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 3.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 1.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.8 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.3 | 3.8 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.3 | 4.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 2.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 9.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 1.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 3.8 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 1.3 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.3 | 2.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 1.3 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.2 | 1.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 1.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 6.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 5.0 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.2 | 6.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 6.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 2.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.2 | 5.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 1.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 0.9 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 3.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 17.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 3.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 1.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 2.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 23.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 1.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 0.8 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 1.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 3.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 16.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 1.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 2.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 8.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 22.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 4.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 1.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 6.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 1.0 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 7.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 2.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 9.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 1.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 13.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 7.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 3.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 1.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.3 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 0.1 | 1.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.9 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.9 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 2.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 3.0 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 4.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 4.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 3.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 2.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 30.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 6.3 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.1 | 0.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 2.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 3.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 3.0 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 3.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 3.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.2 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 1.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.6 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 4.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 6.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 1.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 7.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 10.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 4.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 1.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 1.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.5 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 2.9 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.5 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 1.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.7 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.0 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 5.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 51.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.4 | 11.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 3.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.3 | 2.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.3 | 9.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 16.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 4.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 24.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 1.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.3 | 4.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 6.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 5.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 3.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 9.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 10.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 4.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 10.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 6.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 16.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 5.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 6.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.9 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 0.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 5.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 2.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 4.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 32.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.3 | 26.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.7 | 25.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.6 | 6.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.6 | 4.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.6 | 4.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.5 | 5.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.5 | 7.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 29.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 13.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 5.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 1.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 7.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.3 | 5.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 4.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 14.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 6.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 2.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 4.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.3 | 4.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 3.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 3.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 7.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 3.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 7.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 5.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 19.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 5.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.2 | 1.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 10.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 8.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 1.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 2.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 3.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 5.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 6.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 11.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 3.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 11.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.8 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 2.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 5.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 5.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 5.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.5 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.1 | 1.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.0 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 1.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 1.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 3.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 8.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 2.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 3.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.5 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |