Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nanog
Z-value: 0.97
Transcription factors associated with Nanog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nanog
|
ENSMUSG00000012396.13 | Nanog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nanog | mm39_v1_chr6_+_122684448_122684560 | -0.18 | 1.4e-01 | Click! |
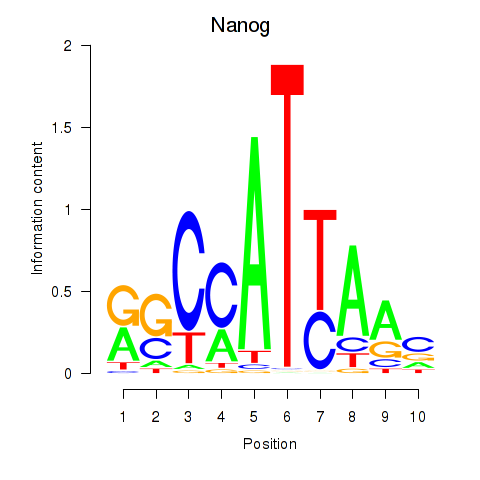
Activity profile of Nanog motif
Sorted Z-values of Nanog motif
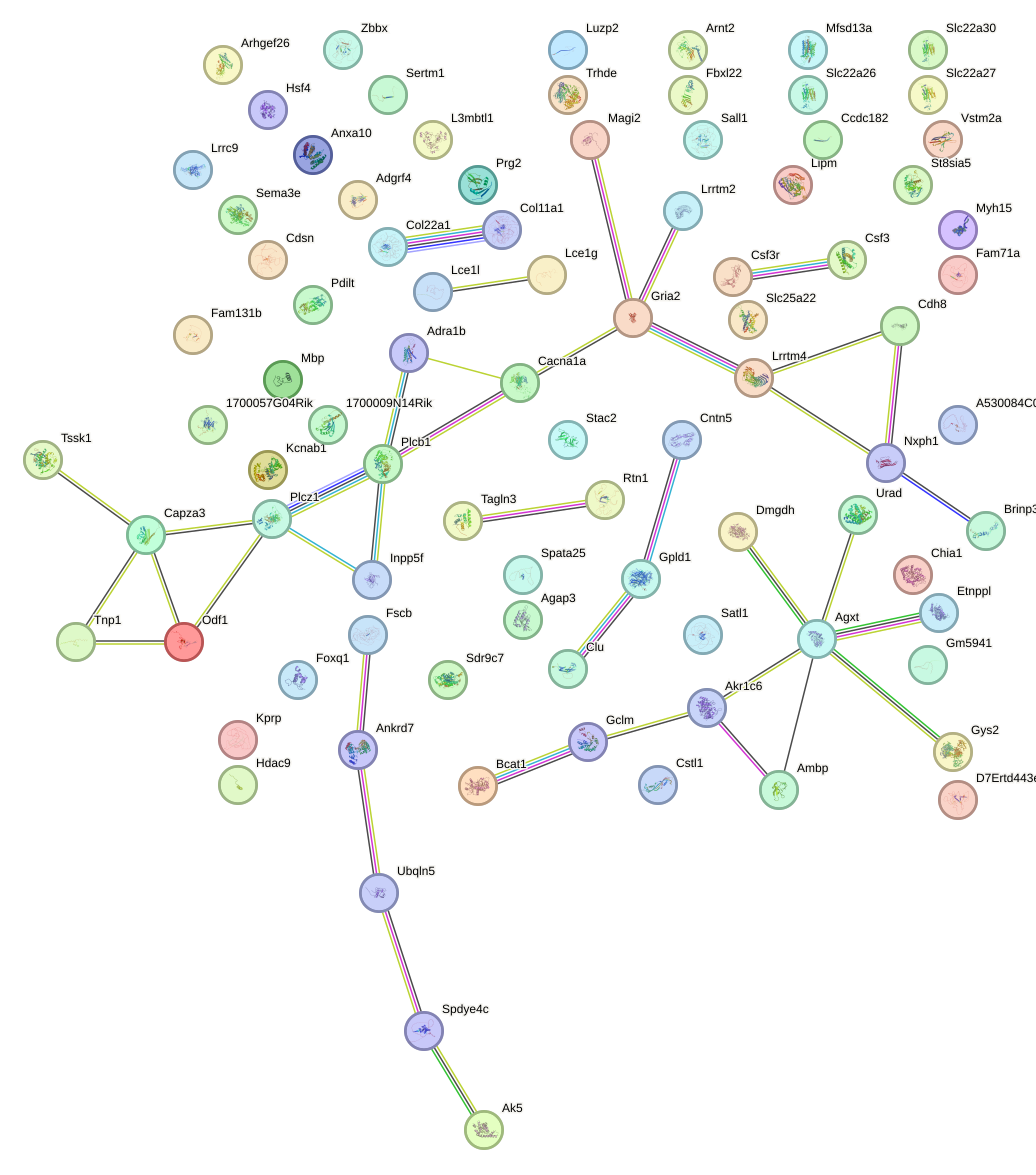
Network of associatons between targets according to the STRING database.
First level regulatory network of Nanog
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_130411097 | 3.76 |
ENSMUST00000166187.8
ENSMUST00000072271.13 |
Etnppl
|
ethanolamine phosphate phospholyase |
| chr3_-_72965136 | 3.60 |
ENSMUST00000059407.9
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr17_+_46608333 | 3.37 |
ENSMUST00000188223.7
ENSMUST00000061722.13 ENSMUST00000166280.8 |
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr18_-_35348049 | 3.21 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr10_+_127734384 | 3.17 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr1_+_146373352 | 3.11 |
ENSMUST00000132847.8
ENSMUST00000166814.8 |
Brinp3
|
bone morphogenetic protein/retinoic acid inducible neural specific 3 |
| chr6_-_144993451 | 3.07 |
ENSMUST00000123930.8
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr7_-_103778992 | 2.92 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr7_+_54485336 | 2.89 |
ENSMUST00000082373.8
|
Luzp2
|
leucine zipper protein 2 |
| chr13_+_93810911 | 2.84 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr8_+_10299288 | 2.83 |
ENSMUST00000214643.2
|
Myo16
|
myosin XVI |
| chr1_+_93062962 | 2.81 |
ENSMUST00000027491.7
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr15_+_21111428 | 2.79 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr3_+_130411294 | 2.77 |
ENSMUST00000163620.8
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr6_+_79995860 | 2.69 |
ENSMUST00000147663.8
ENSMUST00000128718.8 ENSMUST00000126005.8 ENSMUST00000133918.8 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr6_-_144993362 | 2.60 |
ENSMUST00000149769.6
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr18_+_82572595 | 2.60 |
ENSMUST00000152071.9
ENSMUST00000142850.9 ENSMUST00000080658.12 ENSMUST00000133193.9 ENSMUST00000123251.9 ENSMUST00000153478.9 ENSMUST00000075372.13 ENSMUST00000102812.12 ENSMUST00000062446.15 ENSMUST00000114674.11 ENSMUST00000132369.3 |
Mbp
|
myelin basic protein |
| chr1_-_73055043 | 2.54 |
ENSMUST00000027374.7
|
Tnp1
|
transition protein 1 |
| chr15_+_38219447 | 2.48 |
ENSMUST00000081966.5
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr14_+_66208059 | 2.41 |
ENSMUST00000127387.8
|
Clu
|
clusterin |
| chr8_-_100143029 | 2.32 |
ENSMUST00000155527.8
ENSMUST00000142129.8 ENSMUST00000093249.11 ENSMUST00000142475.3 ENSMUST00000128860.8 |
Cdh8
|
cadherin 8 |
| chr13_+_4486105 | 2.29 |
ENSMUST00000156277.2
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr7_+_128290204 | 2.29 |
ENSMUST00000118605.2
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr4_-_63072367 | 2.24 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr7_-_119122681 | 2.19 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr6_+_79995994 | 2.17 |
ENSMUST00000126399.2
ENSMUST00000136421.2 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr19_-_8382424 | 2.17 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr6_-_139987135 | 2.16 |
ENSMUST00000032356.13
|
Plcz1
|
phospholipase C, zeta 1 |
| chr11_+_16207705 | 2.15 |
ENSMUST00000109645.9
ENSMUST00000109647.3 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr14_+_66208253 | 2.08 |
ENSMUST00000138191.8
|
Clu
|
clusterin |
| chr12_-_72455708 | 2.08 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr8_-_89770790 | 2.06 |
ENSMUST00000034090.8
|
Sall1
|
spalt like transcription factor 1 |
| chr13_+_25127127 | 1.97 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr3_+_113824181 | 1.96 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr16_-_45544960 | 1.93 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr3_-_92734546 | 1.90 |
ENSMUST00000072363.5
|
Kprp
|
keratinocyte expressed, proline-rich |
| chr12_+_72488625 | 1.90 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr10_+_90412432 | 1.88 |
ENSMUST00000182786.8
ENSMUST00000182600.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr7_+_46636562 | 1.88 |
ENSMUST00000185832.2
|
Gm9999
|
predicted gene 9999 |
| chr6_-_142418801 | 1.88 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr16_-_9812787 | 1.87 |
ENSMUST00000199708.5
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr8_-_62576140 | 1.85 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr6_+_8948608 | 1.84 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr12_-_34578842 | 1.81 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr3_-_75051076 | 1.81 |
ENSMUST00000107778.9
ENSMUST00000107775.8 |
Zbbx
|
zinc finger, B-box domain containing |
| chr6_+_139987275 | 1.80 |
ENSMUST00000043797.6
|
Capza3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr7_+_6733561 | 1.77 |
ENSMUST00000200535.6
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr8_+_105996469 | 1.77 |
ENSMUST00000172525.8
ENSMUST00000174837.8 ENSMUST00000173859.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr3_-_75051010 | 1.76 |
ENSMUST00000107776.8
ENSMUST00000039269.13 |
Zbbx
|
zinc finger, B-box domain containing |
| chr19_-_7943365 | 1.75 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr3_+_62327089 | 1.75 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr4_+_101365052 | 1.74 |
ENSMUST00000038207.12
|
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr14_+_66208498 | 1.72 |
ENSMUST00000128539.8
|
Clu
|
clusterin |
| chr6_+_18866308 | 1.71 |
ENSMUST00000031489.10
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr6_-_42301574 | 1.70 |
ENSMUST00000031891.15
ENSMUST00000143278.8 |
Fam131b
|
family with sequence similarity 131, member B |
| chr5_+_14075281 | 1.66 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr4_+_101365144 | 1.66 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr2_+_162785394 | 1.66 |
ENSMUST00000035751.12
ENSMUST00000156954.8 |
L3mbtl1
|
L3MBTL1 histone methyl-lysine binding protein |
| chr9_-_10904714 | 1.65 |
ENSMUST00000162484.8
ENSMUST00000160216.8 |
Cntn5
|
contactin 5 |
| chr19_+_34078333 | 1.65 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr5_+_20112704 | 1.65 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_+_24679154 | 1.63 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr3_-_54823287 | 1.60 |
ENSMUST00000070342.4
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr15_-_71826243 | 1.60 |
ENSMUST00000229585.2
|
Col22a1
|
collagen, type XXII, alpha 1 |
| chr17_-_43003135 | 1.59 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr16_-_9812410 | 1.59 |
ENSMUST00000115835.8
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr10_-_114638202 | 1.58 |
ENSMUST00000239411.2
|
Trhde
|
TRH-degrading enzyme |
| chr16_+_17712061 | 1.58 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr2_+_148592272 | 1.58 |
ENSMUST00000109955.10
ENSMUST00000109954.8 ENSMUST00000109952.2 |
Cstl1
|
cystatin-like 1 |
| chr3_-_92758591 | 1.57 |
ENSMUST00000054426.5
|
Lce1l
|
late cornified envelope 1L |
| chr11_-_43792013 | 1.54 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr6_-_42301488 | 1.53 |
ENSMUST00000095974.4
|
Fam131b
|
family with sequence similarity 131, member B |
| chr11_-_97944239 | 1.53 |
ENSMUST00000017544.9
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr12_-_64521464 | 1.51 |
ENSMUST00000059833.8
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr2_+_134627987 | 1.51 |
ENSMUST00000131552.5
ENSMUST00000110116.8 |
Plcb1
|
phospholipase C, beta 1 |
| chr3_-_54962458 | 1.50 |
ENSMUST00000199352.2
ENSMUST00000198320.5 ENSMUST00000029368.7 |
Ccna1
|
cyclin A1 |
| chr3_-_152373997 | 1.48 |
ENSMUST00000045262.11
|
Ak5
|
adenylate kinase 5 |
| chr2_-_164670452 | 1.47 |
ENSMUST00000017911.4
|
Spata25
|
spermatogenesis associated 25 |
| chrX_+_91533555 | 1.47 |
ENSMUST00000096371.3
|
Gm5941
|
predicted gene 5941 |
| chr3_+_106020545 | 1.45 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr7_-_141014445 | 1.45 |
ENSMUST00000133021.2
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr3_+_64884839 | 1.42 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr8_+_85142413 | 1.41 |
ENSMUST00000215756.2
|
Cacna1a
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr7_-_18852282 | 1.40 |
ENSMUST00000141380.3
|
Meiosin
|
meiosis initiator |
| chr7_-_84059170 | 1.40 |
ENSMUST00000208995.2
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr11_+_88184872 | 1.40 |
ENSMUST00000037268.6
|
Ccdc182
|
coiled-coil domain containing 182 |
| chr18_+_77273510 | 1.39 |
ENSMUST00000075290.8
ENSMUST00000079618.11 |
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr9_-_66421868 | 1.39 |
ENSMUST00000056890.10
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr3_-_80710097 | 1.38 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr17_+_3447465 | 1.38 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr7_-_84059321 | 1.36 |
ENSMUST00000085077.5
ENSMUST00000207769.2 |
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr7_-_133943703 | 1.34 |
ENSMUST00000106129.9
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr2_+_128433125 | 1.34 |
ENSMUST00000155430.8
|
Spdye4c
|
speedy/RINGO cell cycle regulator family, member E4C |
| chr4_+_125918333 | 1.34 |
ENSMUST00000106162.8
|
Csf3r
|
colony stimulating factor 3 receptor (granulocyte) |
| chr1_-_190897012 | 1.33 |
ENSMUST00000171798.2
|
Fam71a
|
family with sequence similarity 71, member A |
| chr5_+_20112500 | 1.33 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_+_98592089 | 1.33 |
ENSMUST00000038886.3
|
Csf3
|
colony stimulating factor 3 (granulocyte) |
| chr13_+_31740117 | 1.33 |
ENSMUST00000042118.11
|
Foxq1
|
forkhead box Q1 |
| chr10_-_95678786 | 1.32 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr4_+_39450265 | 1.31 |
ENSMUST00000029955.5
|
1700009N14Rik
|
RIKEN cDNA 1700009N14 gene |
| chr3_+_122039206 | 1.29 |
ENSMUST00000029769.14
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr19_-_7779943 | 1.29 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr5_-_147259245 | 1.27 |
ENSMUST00000100433.5
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
| chr9_+_92191415 | 1.27 |
ENSMUST00000150594.8
ENSMUST00000098477.8 |
1700057G04Rik
|
RIKEN cDNA 1700057G04 gene |
| chrX_-_111316476 | 1.27 |
ENSMUST00000026601.3
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr16_+_48877762 | 1.26 |
ENSMUST00000168680.2
|
Myh15
|
myosin, heavy chain 15 |
| chr7_-_141014477 | 1.26 |
ENSMUST00000106007.10
ENSMUST00000150026.2 ENSMUST00000202840.4 ENSMUST00000133206.9 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr7_+_6733684 | 1.26 |
ENSMUST00000197117.5
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr10_-_95678748 | 1.24 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr19_+_46345319 | 1.24 |
ENSMUST00000086969.13
|
Mfsd13a
|
major facilitator superfamily domain containing 13a |
| chr7_-_141241632 | 1.24 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr5_+_30869193 | 1.24 |
ENSMUST00000088081.11
ENSMUST00000101442.4 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr14_-_40726472 | 1.23 |
ENSMUST00000153830.8
|
Prxl2a
|
peroxiredoxin like 2A |
| chr14_+_10123804 | 1.23 |
ENSMUST00000022262.6
ENSMUST00000224714.2 |
Fezf2
|
Fez family zinc finger 2 |
| chr17_+_35863025 | 1.23 |
ENSMUST00000044804.8
|
Cdsn
|
corneodesmosin |
| chr7_+_106808645 | 1.22 |
ENSMUST00000098135.2
|
Rbmxl2
|
RNA binding motif protein, X-linked-like 2 |
| chr2_+_85551751 | 1.22 |
ENSMUST00000055517.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr14_-_96756503 | 1.22 |
ENSMUST00000022666.9
|
Klhl1
|
kelch-like 1 |
| chr19_-_7183596 | 1.21 |
ENSMUST00000123594.8
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr16_+_20514925 | 1.21 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr5_-_104125226 | 1.21 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr13_+_12580743 | 1.20 |
ENSMUST00000221560.2
ENSMUST00000071973.8 |
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr1_-_132318039 | 1.20 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr3_+_59914164 | 1.19 |
ENSMUST00000169794.2
|
Aadacl2
|
arylacetamide deacetylase like 2 |
| chr19_+_44980565 | 1.17 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr10_+_69621337 | 1.16 |
ENSMUST00000182993.8
|
Ank3
|
ankyrin 3, epithelial |
| chr12_-_76869510 | 1.16 |
ENSMUST00000154765.8
|
Rab15
|
RAB15, member RAS oncogene family |
| chr5_+_36641922 | 1.15 |
ENSMUST00000060100.3
|
Ccdc96
|
coiled-coil domain containing 96 |
| chr7_-_141015240 | 1.15 |
ENSMUST00000138865.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr15_+_79982033 | 1.14 |
ENSMUST00000143928.2
|
Syngr1
|
synaptogyrin 1 |
| chr7_-_85985625 | 1.13 |
ENSMUST00000069279.5
|
Olfr307
|
olfactory receptor 307 |
| chr7_+_16821858 | 1.13 |
ENSMUST00000152671.2
|
Psg16
|
pregnancy specific glycoprotein 16 |
| chr2_+_62494622 | 1.11 |
ENSMUST00000028257.3
|
Gca
|
grancalcin |
| chr11_-_20282684 | 1.11 |
ENSMUST00000004634.7
ENSMUST00000109594.8 |
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr3_-_54962899 | 1.08 |
ENSMUST00000199144.5
|
Ccna1
|
cyclin A1 |
| chr3_-_57599956 | 1.08 |
ENSMUST00000238789.2
ENSMUST00000197088.5 ENSMUST00000099091.4 |
Ankub1
|
ankrin repeat and ubiquitin domain containing 1 |
| chr1_+_172383499 | 1.08 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr19_-_11618192 | 1.06 |
ENSMUST00000112984.4
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr8_+_46944000 | 1.06 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr6_-_102441628 | 1.05 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr19_-_7183626 | 1.04 |
ENSMUST00000025679.11
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr19_-_11618165 | 1.03 |
ENSMUST00000186023.7
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr10_-_128796834 | 1.03 |
ENSMUST00000026398.5
|
Mettl7b
|
methyltransferase like 7B |
| chr9_-_44253630 | 1.02 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr10_-_78554104 | 1.02 |
ENSMUST00000005488.9
|
Casp14
|
caspase 14 |
| chr3_+_138911419 | 1.02 |
ENSMUST00000106239.8
|
Stpg2
|
sperm tail PG rich repeat containing 2 |
| chr5_-_104125270 | 1.02 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr11_+_102775991 | 1.01 |
ENSMUST00000100369.4
|
Fam187a
|
family with sequence similarity 187, member A |
| chrX_+_48559432 | 1.01 |
ENSMUST00000042444.7
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr17_-_90395568 | 1.01 |
ENSMUST00000173222.2
|
Nrxn1
|
neurexin I |
| chr11_-_109188947 | 1.00 |
ENSMUST00000020920.10
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr6_+_41331039 | 1.00 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chrX_+_8137881 | 0.98 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr7_-_141014192 | 0.96 |
ENSMUST00000201127.5
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr15_+_9436114 | 0.95 |
ENSMUST00000042360.5
ENSMUST00000226688.2 |
Capsl
|
calcyphosine-like |
| chrX_+_48559327 | 0.94 |
ENSMUST00000114904.10
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr7_-_25454177 | 0.93 |
ENSMUST00000206832.2
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr12_+_37930169 | 0.93 |
ENSMUST00000221176.2
|
Dgkb
|
diacylglycerol kinase, beta |
| chr5_-_116427003 | 0.92 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr19_+_39499288 | 0.92 |
ENSMUST00000025968.5
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr2_-_139908615 | 0.92 |
ENSMUST00000046656.9
ENSMUST00000099304.4 ENSMUST00000110079.9 |
Tasp1
|
taspase, threonine aspartase 1 |
| chr5_+_81169049 | 0.91 |
ENSMUST00000117253.8
ENSMUST00000120128.8 |
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chrX_+_106836189 | 0.90 |
ENSMUST00000101292.9
|
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr6_+_30610973 | 0.90 |
ENSMUST00000062758.11
|
Cpa5
|
carboxypeptidase A5 |
| chr8_-_55340024 | 0.89 |
ENSMUST00000176866.8
|
Wdr17
|
WD repeat domain 17 |
| chr19_+_6096606 | 0.88 |
ENSMUST00000138532.8
ENSMUST00000129081.8 ENSMUST00000156550.8 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr15_-_79658584 | 0.88 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr2_-_166904902 | 0.88 |
ENSMUST00000048988.14
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chrX_-_111315519 | 0.87 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr7_-_45343785 | 0.87 |
ENSMUST00000040636.9
|
Sec1
|
secretory blood group 1 |
| chr9_+_21746785 | 0.87 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr7_-_141014336 | 0.87 |
ENSMUST00000136354.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr5_+_20112771 | 0.87 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_-_121410152 | 0.87 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chr14_-_63415235 | 0.86 |
ENSMUST00000054963.10
|
Fdft1
|
farnesyl diphosphate farnesyl transferase 1 |
| chr1_+_46464625 | 0.85 |
ENSMUST00000189749.7
|
Dnah7c
|
dynein, axonemal, heavy chain 7C |
| chr15_-_79658608 | 0.84 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr18_+_61820982 | 0.84 |
ENSMUST00000025471.4
|
Il17b
|
interleukin 17B |
| chr1_-_75110511 | 0.84 |
ENSMUST00000027405.6
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr5_+_107655487 | 0.84 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr4_+_41966058 | 0.83 |
ENSMUST00000108026.3
|
Fam205a4
|
family with sequence similarity 205, member A4 |
| chr5_-_36641456 | 0.83 |
ENSMUST00000119916.2
ENSMUST00000031097.8 |
Tada2b
|
transcriptional adaptor 2B |
| chr17_+_41121979 | 0.82 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr3_-_82783790 | 0.82 |
ENSMUST00000048647.14
|
Rbm46
|
RNA binding motif protein 46 |
| chr13_-_23945189 | 0.81 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1 |
| chr1_-_28819331 | 0.81 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr3_-_37366567 | 0.81 |
ENSMUST00000075537.7
ENSMUST00000071400.13 ENSMUST00000102955.11 ENSMUST00000140956.8 |
Cetn4
|
centrin 4 |
| chr3_+_89043440 | 0.81 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr4_-_135714465 | 0.80 |
ENSMUST00000105851.9
|
Pithd1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chr7_-_115445315 | 0.80 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr7_+_101545547 | 0.80 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr9_-_44253588 | 0.78 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr3_-_97318495 | 0.77 |
ENSMUST00000060912.4
|
Olfr1402
|
olfactory receptor 1402 |
| chr10_+_59239466 | 0.77 |
ENSMUST00000009790.14
|
Pla2g12b
|
phospholipase A2, group XIIB |
| chr7_-_26895141 | 0.77 |
ENSMUST00000163311.9
ENSMUST00000126211.2 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr6_+_30611028 | 0.76 |
ENSMUST00000115138.8
|
Cpa5
|
carboxypeptidase A5 |
| chr18_+_37433852 | 0.76 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr4_-_88510622 | 0.75 |
ENSMUST00000102807.2
|
Ifna9
|
interferon alpha 9 |
| chr9_+_19624125 | 0.75 |
ENSMUST00000077023.4
|
Olfr857
|
olfactory receptor 857 |
| chr4_-_88562696 | 0.74 |
ENSMUST00000105149.3
|
Ifna13
|
interferon alpha 13 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.2 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.9 | 2.8 | GO:0042853 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) oxalic acid secretion(GO:0046724) |
| 0.8 | 2.3 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.7 | 5.7 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.7 | 2.6 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.6 | 1.8 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.6 | 3.4 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.5 | 3.8 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.5 | 1.5 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.5 | 2.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.5 | 2.5 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.5 | 2.0 | GO:0006507 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.4 | 3.5 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.4 | 2.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 | 4.8 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.4 | 2.0 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.4 | 1.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.4 | 1.4 | GO:0021679 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) |
| 0.3 | 5.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 2.8 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 1.5 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.3 | 0.9 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.3 | 2.0 | GO:0035989 | tendon development(GO:0035989) |
| 0.3 | 4.9 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.2 | 0.9 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 1.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 0.6 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 0.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 2.0 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.2 | 1.2 | GO:1904936 | forebrain anterior/posterior pattern specification(GO:0021797) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 1.9 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 1.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 5.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 1.7 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 2.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.6 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 1.3 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 3.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 1.5 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 1.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 1.0 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 2.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.2 | GO:0045818 | negative regulation of glycogen catabolic process(GO:0045818) |
| 0.1 | 0.4 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.1 | 1.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.3 | GO:0060450 | positive regulation of hindgut contraction(GO:0060450) |
| 0.1 | 0.2 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 1.1 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.4 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 2.6 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 1.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.8 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.6 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.7 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 1.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.7 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 1.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 2.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.2 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 1.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.3 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 1.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.2 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 1.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.4 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 2.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 | 2.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.8 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.1 | 0.2 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 2.2 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 0.2 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 1.0 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 1.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 | 0.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.9 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.8 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 1.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 2.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 1.7 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 4.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 2.4 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.6 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.6 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 1.6 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.6 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0046084 | adenine salvage(GO:0006168) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 1.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 1.2 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 1.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.8 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 1.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0030953 | astral microtubule organization(GO:0030953) cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 2.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.8 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.5 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.4 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 1.3 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.2 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.3 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 1.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 12.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.0 | GO:0061738 | multivesicular body assembly(GO:0036258) mitotic cytokinesis checkpoint(GO:0044878) late endosomal microautophagy(GO:0061738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.4 | 1.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.4 | 6.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 2.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 4.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 0.7 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.2 | 0.6 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.2 | 1.6 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 2.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 4.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.2 | 2.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 2.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 4.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.5 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.4 | GO:1990037 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.9 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.4 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 1.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 2.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 1.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 5.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.2 | GO:0044438 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 5.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.2 | GO:0000793 | condensed chromosome(GO:0000793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.8 | 2.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.7 | 2.8 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.7 | 5.7 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.7 | 2.8 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.6 | 1.9 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.6 | 1.8 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.6 | 2.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.5 | 2.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.5 | 3.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 2.3 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.4 | 1.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.4 | 1.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.4 | 1.7 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.4 | 1.5 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.4 | 5.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.4 | 1.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.3 | 2.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 5.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 0.9 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.2 | 3.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 6.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 1.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 1.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 2.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 1.1 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.2 | 2.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 2.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 2.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.8 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 1.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 0.8 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 0.9 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 4.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 3.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.7 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 3.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 3.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 1.1 | GO:0015193 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.7 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 1.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.7 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 1.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 2.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 2.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0003999 | adenine binding(GO:0002055) adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.8 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 1.5 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.4 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 14.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.2 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 5.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 2.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 4.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 4.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 3.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 2.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 4.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.3 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.4 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |