Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
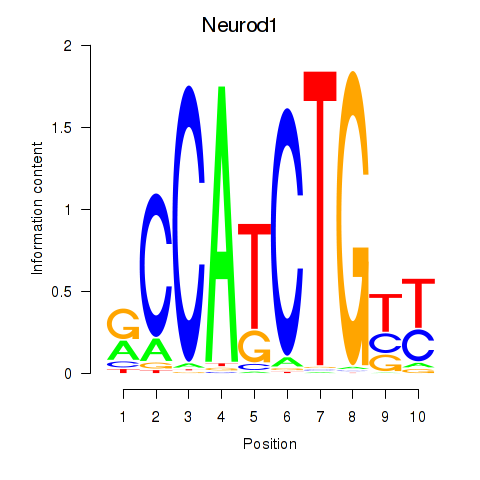
Results for Neurod1
Z-value: 1.38
Transcription factors associated with Neurod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurod1
|
ENSMUSG00000034701.10 | Neurod1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurod1 | mm39_v1_chr2_-_79287095_79287102 | 0.44 | 1.1e-04 | Click! |
Activity profile of Neurod1 motif
Sorted Z-values of Neurod1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurod1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_180310894 | 10.57 |
ENSMUST00000211561.2
ENSMUST00000136521.2 ENSMUST00000179826.2 |
Stum
|
mechanosensory transduction mediator |
| chr1_+_162466717 | 8.93 |
ENSMUST00000028020.11
|
Myoc
|
myocilin |
| chr3_-_126792056 | 8.68 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr10_+_127702326 | 8.41 |
ENSMUST00000092058.4
|
Rdh16f2
|
RDH16 family member 2 |
| chr2_-_25209107 | 8.25 |
ENSMUST00000114318.10
ENSMUST00000114310.10 ENSMUST00000114308.10 ENSMUST00000114317.10 ENSMUST00000028335.13 ENSMUST00000114314.10 ENSMUST00000114307.8 |
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr18_+_45402018 | 7.39 |
ENSMUST00000183850.8
ENSMUST00000066890.14 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr10_+_69542153 | 7.13 |
ENSMUST00000182992.8
|
Ank3
|
ankyrin 3, epithelial |
| chr6_-_55658242 | 6.93 |
ENSMUST00000044767.10
|
Neurod6
|
neurogenic differentiation 6 |
| chr1_+_9671388 | 6.71 |
ENSMUST00000088666.4
|
Vxn
|
vexin |
| chr8_+_45960804 | 6.53 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr5_-_142594549 | 6.34 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr5_-_37146266 | 5.89 |
ENSMUST00000166339.8
|
Wfs1
|
wolframin ER transmembrane glycoprotein |
| chr8_+_23629173 | 5.84 |
ENSMUST00000174435.2
|
Ank1
|
ankyrin 1, erythroid |
| chr16_+_6166982 | 5.74 |
ENSMUST00000056416.9
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_-_10135449 | 5.55 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr8_+_23629046 | 5.14 |
ENSMUST00000121075.8
|
Ank1
|
ankyrin 1, erythroid |
| chr3_-_84167119 | 5.12 |
ENSMUST00000107691.8
|
Trim2
|
tripartite motif-containing 2 |
| chr1_-_79417732 | 5.02 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr18_-_12952925 | 4.98 |
ENSMUST00000119043.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chrX_+_72546680 | 4.96 |
ENSMUST00000033744.12
ENSMUST00000088429.8 ENSMUST00000114479.2 |
Atp2b3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr2_-_79959178 | 4.76 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_-_141015240 | 4.72 |
ENSMUST00000138865.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr2_-_53975501 | 4.70 |
ENSMUST00000100089.3
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr10_-_80649315 | 4.53 |
ENSMUST00000181039.8
ENSMUST00000180438.2 |
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr7_+_19144950 | 4.52 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr6_-_54543446 | 4.46 |
ENSMUST00000019268.11
|
Scrn1
|
secernin 1 |
| chr9_+_43978290 | 4.42 |
ENSMUST00000034508.14
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr9_+_121606750 | 4.40 |
ENSMUST00000098272.4
|
Klhl40
|
kelch-like 40 |
| chr16_+_20408886 | 4.38 |
ENSMUST00000232279.2
ENSMUST00000232474.2 |
Vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr3_+_18108313 | 4.38 |
ENSMUST00000026120.8
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr1_+_143516402 | 4.36 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr1_-_93029532 | 4.35 |
ENSMUST00000171796.8
|
Kif1a
|
kinesin family member 1A |
| chr6_-_124410452 | 4.30 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr1_-_173195236 | 4.27 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr11_-_5900019 | 4.27 |
ENSMUST00000102920.4
|
Gck
|
glucokinase |
| chr4_+_80828883 | 4.25 |
ENSMUST00000055922.4
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr1_-_93029547 | 4.16 |
ENSMUST00000112958.9
ENSMUST00000186861.2 ENSMUST00000171556.8 |
Kif1a
|
kinesin family member 1A |
| chr10_+_3316505 | 4.14 |
ENSMUST00000217573.2
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr8_+_120121612 | 4.12 |
ENSMUST00000098367.5
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr18_-_89787603 | 4.12 |
ENSMUST00000097495.5
|
Dok6
|
docking protein 6 |
| chr14_+_65504151 | 4.04 |
ENSMUST00000169656.3
ENSMUST00000226005.2 |
Fbxo16
|
F-box protein 16 |
| chr2_+_96148418 | 4.04 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr2_+_121125918 | 4.04 |
ENSMUST00000110639.8
|
Map1a
|
microtubule-associated protein 1 A |
| chr4_-_110144676 | 4.02 |
ENSMUST00000106598.8
ENSMUST00000102723.11 ENSMUST00000153906.2 |
Elavl4
|
ELAV like RNA binding protein 4 |
| chr9_+_107814499 | 4.02 |
ENSMUST00000195219.2
|
Camkv
|
CaM kinase-like vesicle-associated |
| chr14_-_28691423 | 4.00 |
ENSMUST00000225985.2
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr14_+_67470884 | 3.97 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr4_-_141351110 | 3.94 |
ENSMUST00000038661.8
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr8_+_23629080 | 3.89 |
ENSMUST00000033947.15
|
Ank1
|
ankyrin 1, erythroid |
| chr5_-_52628825 | 3.89 |
ENSMUST00000198008.5
ENSMUST00000059428.7 |
Ccdc149
|
coiled-coil domain containing 149 |
| chr5_+_90608751 | 3.81 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr5_+_22951015 | 3.79 |
ENSMUST00000197992.2
|
Lhfpl3
|
lipoma HMGIC fusion partner-like 3 |
| chr13_+_46655589 | 3.79 |
ENSMUST00000119341.2
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr2_-_52225763 | 3.79 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr11_+_102727122 | 3.77 |
ENSMUST00000021302.15
ENSMUST00000107072.2 |
Higd1b
|
HIG1 domain family, member 1B |
| chr8_-_34237752 | 3.75 |
ENSMUST00000179364.3
|
Smim18
|
small integral membrane protein 18 |
| chr13_+_46655617 | 3.72 |
ENSMUST00000225824.2
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr17_+_56312672 | 3.71 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr10_-_108846816 | 3.70 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr2_-_84573999 | 3.65 |
ENSMUST00000181711.2
|
Gm19426
|
predicted gene, 19426 |
| chrX_-_23132991 | 3.62 |
ENSMUST00000115316.9
|
Klhl13
|
kelch-like 13 |
| chr9_+_108708939 | 3.61 |
ENSMUST00000192235.2
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chrX_-_74621828 | 3.59 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr11_-_98220466 | 3.57 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr1_-_93029576 | 3.57 |
ENSMUST00000190723.7
|
Kif1a
|
kinesin family member 1A |
| chr16_+_96001650 | 3.57 |
ENSMUST00000048770.16
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein |
| chr2_-_170248421 | 3.57 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr2_-_92222979 | 3.54 |
ENSMUST00000111279.9
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr16_-_67417768 | 3.54 |
ENSMUST00000114292.8
ENSMUST00000120898.8 |
Cadm2
|
cell adhesion molecule 2 |
| chr6_-_21851827 | 3.53 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr17_-_13070780 | 3.51 |
ENSMUST00000162389.2
ENSMUST00000162119.8 ENSMUST00000159223.8 |
Mas1
|
MAS1 oncogene |
| chr1_-_189902868 | 3.48 |
ENSMUST00000177288.4
ENSMUST00000175916.8 |
Prox1
|
prospero homeobox 1 |
| chr2_+_25293140 | 3.46 |
ENSMUST00000154809.8
ENSMUST00000055921.14 ENSMUST00000141567.8 |
Npdc1
|
neural proliferation, differentiation and control 1 |
| chr12_+_29578354 | 3.45 |
ENSMUST00000218583.2
ENSMUST00000049784.17 |
Myt1l
|
myelin transcription factor 1-like |
| chr5_+_125609440 | 3.36 |
ENSMUST00000031446.7
|
Tmem132b
|
transmembrane protein 132B |
| chr15_-_27788693 | 3.34 |
ENSMUST00000226287.2
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr10_-_127724557 | 3.30 |
ENSMUST00000047199.5
|
Rdh7
|
retinol dehydrogenase 7 |
| chr14_+_67470735 | 3.23 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr9_-_66951234 | 3.22 |
ENSMUST00000113690.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr17_-_57394718 | 3.21 |
ENSMUST00000071135.6
|
Tubb4a
|
tubulin, beta 4A class IVA |
| chr9_-_66951114 | 3.19 |
ENSMUST00000113686.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr6_-_97229708 | 3.19 |
ENSMUST00000095655.4
|
Lmod3
|
leiomodin 3 (fetal) |
| chr9_+_65268304 | 3.18 |
ENSMUST00000147185.3
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr3_+_68491487 | 3.17 |
ENSMUST00000182997.3
|
Schip1
|
schwannomin interacting protein 1 |
| chr2_+_91090167 | 3.17 |
ENSMUST00000138470.2
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr9_+_58489523 | 3.16 |
ENSMUST00000177292.8
ENSMUST00000085651.12 ENSMUST00000176557.8 ENSMUST00000114121.11 ENSMUST00000177064.8 |
Nptn
|
neuroplastin |
| chr1_+_167426019 | 3.12 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chr11_-_35871300 | 3.12 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr5_+_91665474 | 3.10 |
ENSMUST00000040576.10
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr14_-_109151590 | 3.06 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr18_-_35348049 | 3.05 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr13_+_52000704 | 2.98 |
ENSMUST00000021903.3
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr8_+_45960855 | 2.98 |
ENSMUST00000141039.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chrX_+_139243012 | 2.94 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr17_-_56424577 | 2.91 |
ENSMUST00000019808.12
|
Plin5
|
perilipin 5 |
| chr4_-_22488296 | 2.89 |
ENSMUST00000178174.3
|
Pou3f2
|
POU domain, class 3, transcription factor 2 |
| chr15_-_77037972 | 2.84 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr18_+_69654572 | 2.83 |
ENSMUST00000200862.4
|
Tcf4
|
transcription factor 4 |
| chr2_+_90716204 | 2.80 |
ENSMUST00000111466.3
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr11_+_78394273 | 2.80 |
ENSMUST00000001130.8
ENSMUST00000125670.3 |
Sebox
|
SEBOX homeobox |
| chr5_-_49682106 | 2.79 |
ENSMUST00000176191.8
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr2_-_120370333 | 2.78 |
ENSMUST00000171215.8
|
Zfp106
|
zinc finger protein 106 |
| chr2_-_25471703 | 2.75 |
ENSMUST00000114217.3
ENSMUST00000191602.2 |
Ajm1
|
apical junction component 1 |
| chr7_+_16609227 | 2.73 |
ENSMUST00000108493.3
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr2_+_85551751 | 2.73 |
ENSMUST00000055517.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr3_-_73615732 | 2.72 |
ENSMUST00000029367.6
|
Bche
|
butyrylcholinesterase |
| chr14_-_78970160 | 2.67 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr11_+_50917831 | 2.66 |
ENSMUST00000072152.2
|
Olfr54
|
olfactory receptor 54 |
| chr18_-_62044871 | 2.66 |
ENSMUST00000166783.3
ENSMUST00000049378.15 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr10_+_79978127 | 2.66 |
ENSMUST00000003156.15
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr17_-_56424265 | 2.63 |
ENSMUST00000113072.3
|
Plin5
|
perilipin 5 |
| chr10_+_107107558 | 2.61 |
ENSMUST00000105280.5
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr3_-_107851021 | 2.61 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr11_-_94864273 | 2.58 |
ENSMUST00000100551.11
ENSMUST00000152042.2 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr16_+_84571011 | 2.51 |
ENSMUST00000114195.8
|
Jam2
|
junction adhesion molecule 2 |
| chr15_+_30172716 | 2.49 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr18_+_69654992 | 2.47 |
ENSMUST00000201627.4
|
Tcf4
|
transcription factor 4 |
| chr11_+_67689094 | 2.47 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr11_-_76468527 | 2.47 |
ENSMUST00000176179.8
|
Abr
|
active BCR-related gene |
| chr17_-_24752683 | 2.45 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr15_-_77037756 | 2.45 |
ENSMUST00000227314.2
ENSMUST00000227930.2 ENSMUST00000227533.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chrX_+_133305529 | 2.43 |
ENSMUST00000113224.9
ENSMUST00000113226.2 |
Drp2
|
dystrophin related protein 2 |
| chr3_-_53771185 | 2.40 |
ENSMUST00000122330.2
ENSMUST00000146598.8 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr18_+_69654231 | 2.39 |
ENSMUST00000202350.4
ENSMUST00000202477.4 |
Tcf4
|
transcription factor 4 |
| chr7_-_64806164 | 2.39 |
ENSMUST00000148459.3
ENSMUST00000119118.8 |
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr14_+_65504067 | 2.37 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr13_+_23728222 | 2.37 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chr7_+_130247912 | 2.36 |
ENSMUST00000207549.2
ENSMUST00000209108.2 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr8_+_127790772 | 2.35 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr11_+_69920956 | 2.34 |
ENSMUST00000232115.2
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chrX_+_133305291 | 2.33 |
ENSMUST00000113228.8
ENSMUST00000153424.8 |
Drp2
|
dystrophin related protein 2 |
| chr4_+_102617332 | 2.32 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr11_+_69920542 | 2.31 |
ENSMUST00000232266.2
ENSMUST00000132597.5 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr11_-_74238498 | 2.31 |
ENSMUST00000080365.6
|
Olfr411
|
olfactory receptor 411 |
| chr10_+_127612243 | 2.30 |
ENSMUST00000136223.2
ENSMUST00000052652.7 |
Rdh9
|
retinol dehydrogenase 9 |
| chr10_+_79978152 | 2.28 |
ENSMUST00000105366.2
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr2_-_13016570 | 2.28 |
ENSMUST00000061545.7
|
C1ql3
|
C1q-like 3 |
| chr15_+_102412157 | 2.27 |
ENSMUST00000096145.5
|
Gm10337
|
predicted gene 10337 |
| chr10_+_127595639 | 2.26 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr19_-_42190589 | 2.26 |
ENSMUST00000018966.8
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr3_-_33136153 | 2.26 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr4_-_110149916 | 2.26 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr15_-_78428865 | 2.24 |
ENSMUST00000053239.4
|
Sstr3
|
somatostatin receptor 3 |
| chr8_-_120362291 | 2.23 |
ENSMUST00000061828.10
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr9_+_43978369 | 2.22 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr8_+_55003359 | 2.21 |
ENSMUST00000033918.4
|
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr2_-_52225146 | 2.20 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr15_-_99603371 | 2.19 |
ENSMUST00000163472.3
|
Gm17349
|
predicted gene, 17349 |
| chr18_+_69654900 | 2.18 |
ENSMUST00000202057.4
|
Tcf4
|
transcription factor 4 |
| chr1_+_167425953 | 2.18 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr7_+_27828855 | 2.16 |
ENSMUST00000059886.12
|
9530053A07Rik
|
RIKEN cDNA 9530053A07 gene |
| chr17_+_46565116 | 2.16 |
ENSMUST00000095262.6
|
Lrrc73
|
leucine rich repeat containing 73 |
| chr13_+_55097200 | 2.15 |
ENSMUST00000026994.14
ENSMUST00000109994.9 |
Unc5a
|
unc-5 netrin receptor A |
| chr10_+_59942020 | 2.13 |
ENSMUST00000121820.9
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr5_-_49682150 | 2.10 |
ENSMUST00000087395.11
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr10_+_127637015 | 2.09 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr5_+_150218847 | 2.09 |
ENSMUST00000239118.2
|
Fry
|
FRY microtubule binding protein |
| chr6_-_93890520 | 2.05 |
ENSMUST00000203688.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr6_-_112466780 | 2.04 |
ENSMUST00000053306.8
|
Oxtr
|
oxytocin receptor |
| chr2_+_81883566 | 2.02 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr5_-_151051000 | 2.02 |
ENSMUST00000202111.4
|
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr11_-_107805830 | 2.02 |
ENSMUST00000039071.3
|
Cacng5
|
calcium channel, voltage-dependent, gamma subunit 5 |
| chr14_+_70768257 | 2.02 |
ENSMUST00000047331.8
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr14_+_70768289 | 2.01 |
ENSMUST00000226548.2
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr17_+_49239393 | 1.99 |
ENSMUST00000046254.3
|
Lrfn2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr7_+_6463510 | 1.99 |
ENSMUST00000056120.5
|
Olfr1336
|
olfactory receptor 1336 |
| chr4_-_152122891 | 1.98 |
ENSMUST00000030792.2
|
Tas1r1
|
taste receptor, type 1, member 1 |
| chr14_+_75693396 | 1.97 |
ENSMUST00000164848.3
|
Siah3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr2_-_79959802 | 1.93 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr14_+_123897383 | 1.89 |
ENSMUST00000049681.14
|
Itgbl1
|
integrin, beta-like 1 |
| chrX_-_141089165 | 1.87 |
ENSMUST00000134825.3
|
Kcne1l
|
potassium voltage-gated channel, Isk-related family, member 1-like, pseudogene |
| chr7_+_30157704 | 1.86 |
ENSMUST00000126297.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr8_-_72124359 | 1.86 |
ENSMUST00000177517.8
ENSMUST00000030170.15 |
Unc13a
|
unc-13 homolog A |
| chr4_+_118522716 | 1.84 |
ENSMUST00000102666.5
|
Olfr62
|
olfactory receptor 62 |
| chr2_-_76812799 | 1.84 |
ENSMUST00000011934.13
ENSMUST00000099981.10 ENSMUST00000099980.10 ENSMUST00000111882.9 ENSMUST00000140091.8 |
Ttn
|
titin |
| chr7_-_30156826 | 1.83 |
ENSMUST00000045817.14
|
Kirrel2
|
kirre like nephrin family adhesion molecule 2 |
| chr1_+_63312420 | 1.82 |
ENSMUST00000239483.2
ENSMUST00000114132.8 ENSMUST00000126932.2 |
Zdbf2
|
zinc finger, DBF-type containing 2 |
| chr1_-_54233207 | 1.82 |
ENSMUST00000120904.8
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_-_75195127 | 1.81 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr1_-_97904958 | 1.81 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr7_+_27307422 | 1.76 |
ENSMUST00000142365.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr14_-_70855980 | 1.75 |
ENSMUST00000228001.2
|
Dmtn
|
dematin actin binding protein |
| chr6_+_145879839 | 1.75 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr13_-_21722197 | 1.75 |
ENSMUST00000168629.2
ENSMUST00000218154.2 |
Olfr1366
|
olfactory receptor 1366 |
| chr5_-_74838461 | 1.73 |
ENSMUST00000117525.8
ENSMUST00000113531.9 ENSMUST00000039744.13 ENSMUST00000121690.8 |
Lnx1
|
ligand of numb-protein X 1 |
| chr5_+_115568638 | 1.72 |
ENSMUST00000131079.8
|
Msi1
|
musashi RNA-binding protein 1 |
| chr8_-_107064615 | 1.71 |
ENSMUST00000067512.8
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr7_-_126046814 | 1.70 |
ENSMUST00000146973.2
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr11_+_32592707 | 1.69 |
ENSMUST00000109366.8
ENSMUST00000093205.13 ENSMUST00000076383.8 |
Fbxw11
|
F-box and WD-40 domain protein 11 |
| chr10_+_78870557 | 1.67 |
ENSMUST00000082244.3
|
Olfr57
|
olfactory receptor 57 |
| chr7_+_3339059 | 1.67 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_-_4973960 | 1.65 |
ENSMUST00000144863.8
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr14_+_50722917 | 1.65 |
ENSMUST00000071932.5
|
Olfr741
|
olfactory receptor 741 |
| chrX_+_36059274 | 1.64 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr6_-_77956635 | 1.64 |
ENSMUST00000161846.8
ENSMUST00000160894.8 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr17_-_56312555 | 1.63 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr3_+_135144202 | 1.63 |
ENSMUST00000166033.6
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_84832783 | 1.63 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr18_-_77652820 | 1.63 |
ENSMUST00000026494.14
ENSMUST00000182024.2 |
Rnf165
|
ring finger protein 165 |
| chr3_-_73615535 | 1.63 |
ENSMUST00000138216.8
|
Bche
|
butyrylcholinesterase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 2.2 | 8.9 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 2.0 | 5.9 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 1.8 | 5.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 1.4 | 1.4 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.4 | 4.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 1.2 | 3.5 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 1.1 | 5.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 1.1 | 6.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.0 | 8.7 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.9 | 3.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.9 | 4.3 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.8 | 3.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.8 | 2.3 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.7 | 4.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.7 | 3.6 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.7 | 5.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.7 | 7.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.6 | 14.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.6 | 2.3 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.6 | 5.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.5 | 7.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.5 | 2.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.5 | 1.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.5 | 3.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 4.4 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.5 | 4.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 1.5 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.5 | 4.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 6.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.5 | 1.4 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.4 | 6.2 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.4 | 2.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 2.6 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.4 | 11.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 3.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.4 | 2.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.4 | 3.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.4 | 2.0 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.4 | 2.0 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.4 | 1.9 | GO:0099624 | atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.4 | 1.5 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.4 | 1.4 | GO:2000292 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.4 | 4.3 | GO:1902474 | regulation of protein localization to synapse(GO:1902473) positive regulation of protein localization to synapse(GO:1902474) |
| 0.4 | 16.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.3 | 2.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.3 | 1.0 | GO:0009946 | proximal/distal axis specification(GO:0009946) bronchiole development(GO:0060435) |
| 0.3 | 5.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.3 | 8.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.3 | 2.0 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.3 | 1.6 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 3.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.3 | 1.9 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.3 | 1.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 2.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.3 | 6.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 0.9 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.3 | 1.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.3 | 2.7 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.3 | 0.9 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.3 | 1.8 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.3 | 1.7 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.3 | 2.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.3 | 8.4 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 7.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.3 | 6.6 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.3 | 1.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 | 2.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 1.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 2.5 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.2 | 1.2 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.2 | 1.8 | GO:0031179 | peptide modification(GO:0031179) |
| 0.2 | 0.9 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 1.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 2.9 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 4.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 1.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 0.7 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.2 | 3.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 5.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.2 | 2.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 3.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 0.6 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.2 | 1.6 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 | 0.7 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.2 | 0.6 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 5.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 4.7 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 1.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 3.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 4.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.7 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 2.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 5.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 6.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.7 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 1.5 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 7.0 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.7 | GO:0009624 | response to nematode(GO:0009624) |
| 0.1 | 6.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 5.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 2.5 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 1.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.7 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 4.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 7.5 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 2.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 7.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 3.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 3.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.7 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 3.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 3.5 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 2.5 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 1.9 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.1 | 1.7 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.9 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.4 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 2.0 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.4 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 1.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 4.6 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.1 | 0.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.8 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 3.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 2.8 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 2.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.2 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.1 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 2.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 2.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.4 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 1.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.4 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.3 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 1.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 3.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.0 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 3.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 7.0 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 7.7 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 2.3 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 3.2 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 1.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.5 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.8 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 2.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 36.0 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 2.7 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.4 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.4 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 1.6 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.2 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 1.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.9 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 3.9 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.5 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 3.9 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 1.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.1 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 1.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 3.2 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.6 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.3 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 1.2 | 3.5 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.8 | 23.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.8 | 2.5 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.7 | 5.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 4.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.5 | 8.9 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.5 | 2.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.5 | 1.5 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.5 | 1.4 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.4 | 8.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 6.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 1.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 1.6 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.3 | 1.9 | GO:0098831 | calyx of Held(GO:0044305) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.3 | 6.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 13.1 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 2.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 2.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.3 | 1.4 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.3 | 4.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 4.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 3.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 4.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 1.7 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 7.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 3.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 0.9 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.2 | 8.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 1.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 3.9 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 3.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 12.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 4.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 6.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 2.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 2.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 10.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 4.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.2 | GO:1990435 | stereocilia tip link(GO:0002140) myosin VII complex(GO:0031477) stereocilia tip-link density(GO:1990427) upper tip-link density(GO:1990435) |
| 0.1 | 11.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.5 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 28.4 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 0.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 6.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 12.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 5.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 2.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 3.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 6.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 7.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 3.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 4.2 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 1.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 6.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 9.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 14.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.4 | 4.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.3 | 8.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.2 | 3.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.1 | 4.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.0 | 8.2 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.9 | 6.2 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.8 | 7.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.8 | 2.5 | GO:0019002 | GMP binding(GO:0019002) |
| 0.7 | 11.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 5.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.6 | 4.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 3.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.6 | 6.7 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.5 | 5.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.5 | 1.6 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.5 | 12.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.5 | 1.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 3.5 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.4 | 4.3 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.4 | 5.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.4 | 1.5 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.4 | 2.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 2.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 2.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.4 | 1.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) |
| 0.3 | 2.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 3.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.3 | 3.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.3 | 8.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 2.0 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 4.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 1.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 0.9 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.3 | 4.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.3 | 1.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 2.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 4.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 3.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 1.5 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 5.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 19.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 3.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 5.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 3.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 3.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 3.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 3.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 2.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 4.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 1.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 6.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 1.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 2.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.8 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 4.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 1.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 6.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 3.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 3.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 7.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.6 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 2.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 2.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.9 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.4 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 1.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.8 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.7 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 4.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 4.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 8.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.5 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 1.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 5.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.0 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 6.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 4.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 5.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 5.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 5.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 6.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 4.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 4.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 36.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 2.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 3.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 4.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.1 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 5.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 9.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 7.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 3.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 3.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 5.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 3.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 7.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 6.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 22.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 3.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 11.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 6.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 3.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 5.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 8.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 4.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 2.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 13.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 6.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 2.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 4.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 10.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 3.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 5.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 4.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 3.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 5.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 2.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 3.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 3.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |