Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
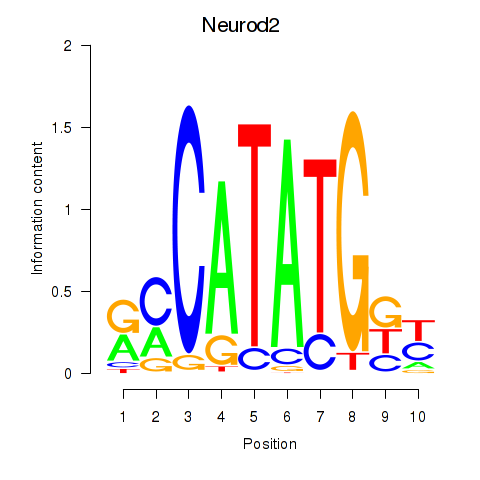
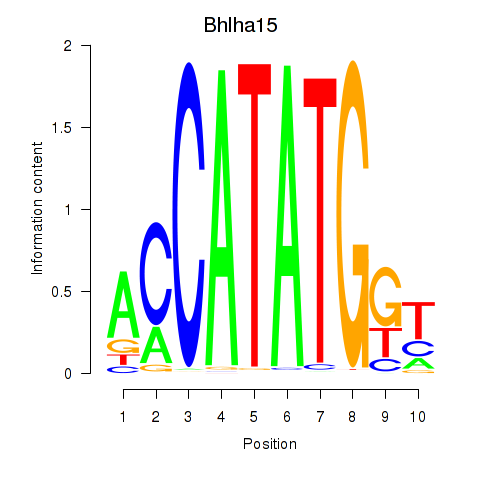
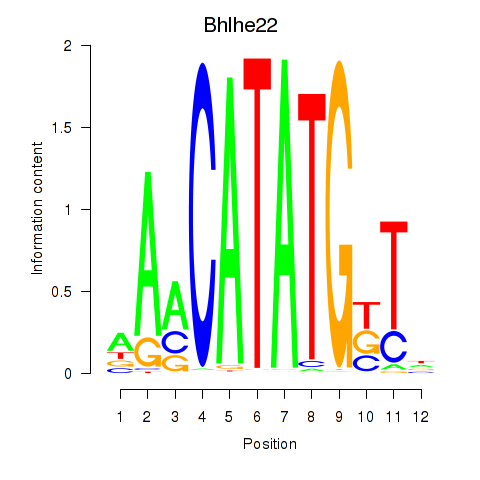
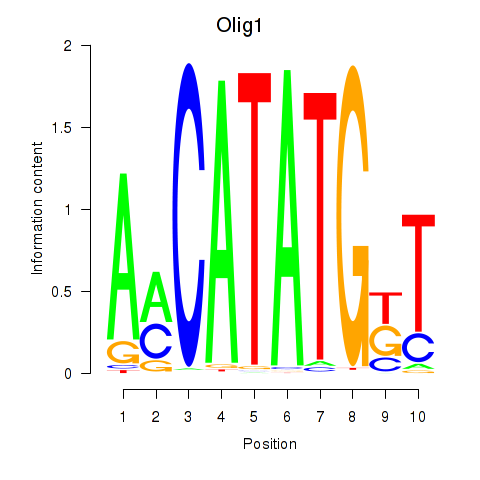
Results for Neurod2_Bhlha15_Bhlhe22_Olig1
Z-value: 0.98
Transcription factors associated with Neurod2_Bhlha15_Bhlhe22_Olig1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurod2
|
ENSMUSG00000038255.7 | Neurod2 |
|
Bhlha15
|
ENSMUSG00000052271.8 | Bhlha15 |
|
Bhlhe22
|
ENSMUSG00000025128.8 | Bhlhe22 |
|
Olig1
|
ENSMUSG00000046160.7 | Olig1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bhlhe22 | mm39_v1_chr3_+_18108313_18108338 | -0.14 | 2.3e-01 | Click! |
| Neurod2 | mm39_v1_chr11_-_98220466_98220482 | -0.12 | 3.3e-01 | Click! |
| Bhlha15 | mm39_v1_chr5_+_144127102_144127152 | -0.10 | 4.0e-01 | Click! |
| Olig1 | mm39_v1_chr16_+_91066602_91066667 | 0.01 | 9.0e-01 | Click! |
Activity profile of Neurod2_Bhlha15_Bhlhe22_Olig1 motif
Sorted Z-values of Neurod2_Bhlha15_Bhlhe22_Olig1 motif
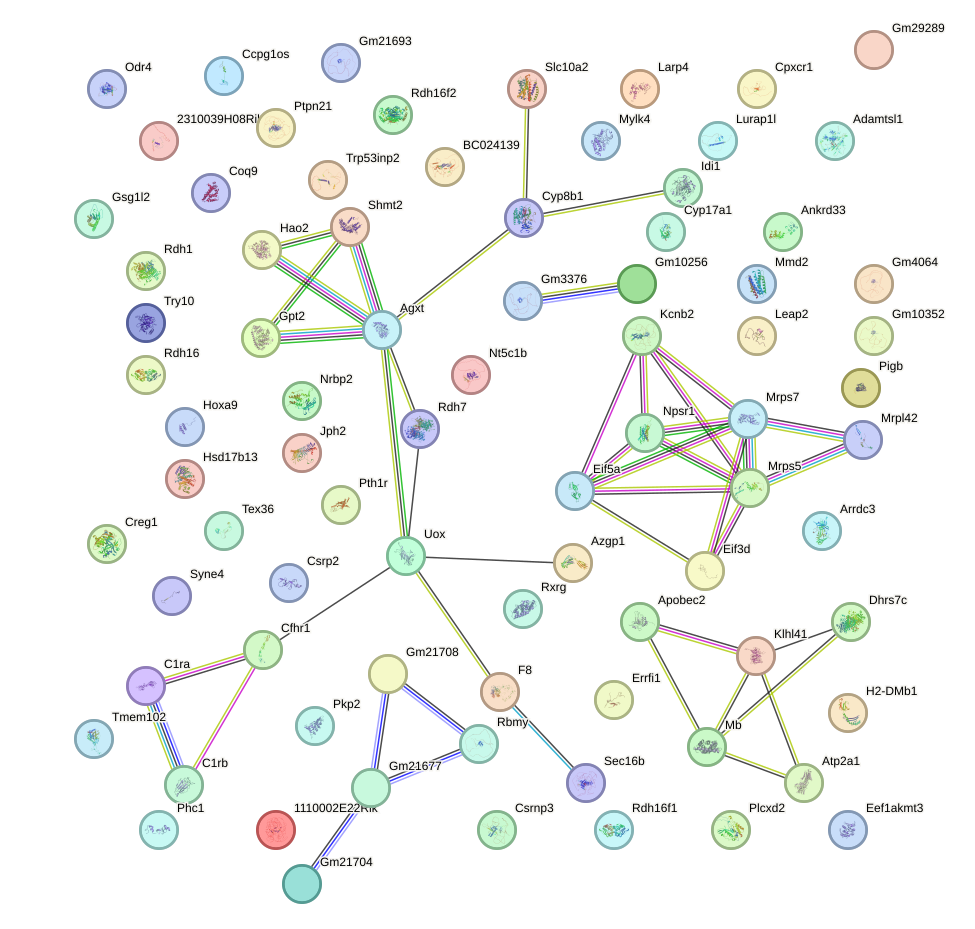
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurod2_Bhlha15_Bhlhe22_Olig1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_16031182 | 12.24 |
ENSMUST00000039408.3
|
Pkp2
|
plakophilin 2 |
| chr8_+_86219191 | 11.92 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr5_-_104125192 | 11.87 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_104125270 | 11.84 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr10_+_87697155 | 10.78 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr5_-_104125226 | 10.49 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr6_+_90439596 | 9.49 |
ENSMUST00000203039.3
|
Klf15
|
Kruppel-like factor 15 |
| chr6_+_90439544 | 9.42 |
ENSMUST00000032174.12
|
Klf15
|
Kruppel-like factor 15 |
| chr2_+_155224105 | 7.39 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr8_+_95564949 | 7.05 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr10_-_127724557 | 6.62 |
ENSMUST00000047199.5
|
Rdh7
|
retinol dehydrogenase 7 |
| chr6_+_124489364 | 5.34 |
ENSMUST00000068593.9
|
C1ra
|
complement component 1, r subcomponent A |
| chr3_+_146302832 | 5.31 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr3_+_137770813 | 5.21 |
ENSMUST00000163080.3
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr5_+_137979763 | 5.20 |
ENSMUST00000035390.7
|
Azgp1
|
alpha-2-glycoprotein 1, zinc |
| chr10_+_127702326 | 5.16 |
ENSMUST00000092058.4
|
Rdh16f2
|
RDH16 family member 2 |
| chr6_-_52203146 | 4.96 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr1_+_93062962 | 4.63 |
ENSMUST00000027491.7
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr9_-_110571645 | 4.36 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr1_-_150268470 | 4.07 |
ENSMUST00000006167.13
ENSMUST00000097547.10 |
Odr4
|
odr4 GPCR localization factor homolog |
| chr1_-_139487951 | 3.84 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr11_-_53313950 | 3.83 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr8_-_5155347 | 3.81 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr2_+_69500444 | 3.73 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chrX_-_74423647 | 3.71 |
ENSMUST00000114085.9
|
F8
|
coagulation factor VIII |
| chr16_+_43056218 | 3.71 |
ENSMUST00000146708.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_+_127637015 | 3.25 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr3_-_27764571 | 3.13 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr9_+_66853343 | 3.00 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr11_-_69696428 | 2.99 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr7_-_126062272 | 2.97 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr9_-_72892617 | 2.96 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr15_-_76010736 | 2.96 |
ENSMUST00000054022.12
ENSMUST00000089654.4 |
BC024139
|
cDNA sequence BC024139 |
| chr15_+_99870714 | 2.73 |
ENSMUST00000230956.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr18_+_12776358 | 2.71 |
ENSMUST00000234966.2
ENSMUST00000025294.9 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr17_+_47083561 | 2.63 |
ENSMUST00000071430.7
|
2310039H08Rik
|
RIKEN cDNA 2310039H08 gene |
| chr10_+_127595639 | 2.56 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr19_-_37153436 | 2.50 |
ENSMUST00000142973.2
ENSMUST00000154376.8 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr9_-_121745354 | 2.38 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr15_-_75963446 | 2.35 |
ENSMUST00000228366.3
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr15_+_99870787 | 2.34 |
ENSMUST00000231160.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr3_-_146302343 | 2.30 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr3_-_98800524 | 2.26 |
ENSMUST00000029464.9
|
Hao2
|
hydroxyacid oxidase 2 |
| chr3_-_27764522 | 2.23 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr1_+_165591315 | 2.18 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr10_-_126877382 | 1.96 |
ENSMUST00000116231.4
|
Eef1akmt3
|
EEF1A lysine methyltransferase 3 |
| chr6_+_41331039 | 1.96 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chrX_+_115358631 | 1.95 |
ENSMUST00000101269.2
|
Cpxcr1
|
CPX chromosome region, candidate 1 |
| chr4_+_80828883 | 1.90 |
ENSMUST00000055922.4
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr12_-_98700886 | 1.84 |
ENSMUST00000085116.4
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr1_+_167445815 | 1.77 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr11_+_67665434 | 1.76 |
ENSMUST00000181566.2
|
Gsg1l2
|
GSG1-like 2 |
| chr11_-_69811717 | 1.70 |
ENSMUST00000152589.2
ENSMUST00000108612.8 ENSMUST00000108611.8 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr9_+_37313287 | 1.67 |
ENSMUST00000115048.10
ENSMUST00000115046.9 ENSMUST00000102895.7 ENSMUST00000239486.2 |
Robo4
|
roundabout guidance receptor 4 |
| chrY_+_2862139 | 1.61 |
ENSMUST00000189964.7
ENSMUST00000188114.2 |
Gm10256
|
predicted gene 10256 |
| chrY_+_3771673 | 1.61 |
ENSMUST00000186140.7
|
Gm3376
|
predicted gene 3376 |
| chrY_-_2796205 | 1.60 |
ENSMUST00000187482.2
|
Gm4064
|
predicted gene 4064 |
| chrY_+_2900989 | 1.60 |
ENSMUST00000187842.7
|
Gm10352
|
predicted gene 10352 |
| chrY_-_3345329 | 1.59 |
ENSMUST00000186047.7
|
Gm21693
|
predicted gene, 21693 |
| chrY_-_3306449 | 1.57 |
ENSMUST00000189592.7
|
Gm21677
|
predicted gene, 21677 |
| chrY_-_3378783 | 1.57 |
ENSMUST00000187277.7
|
Gm21704
|
predicted gene, 21704 |
| chr9_+_37313193 | 1.57 |
ENSMUST00000214185.3
|
Robo4
|
roundabout guidance receptor 4 |
| chrY_+_2830680 | 1.57 |
ENSMUST00000171534.8
ENSMUST00000100360.5 ENSMUST00000179404.8 |
Rbmy
Gm10256
|
RNA binding motif protein, Y chromosome predicted gene 10256 |
| chrY_+_2932582 | 1.55 |
ENSMUST00000188358.2
|
Gm29289
|
predicted gene 29289 |
| chrY_-_3410148 | 1.53 |
ENSMUST00000190283.7
ENSMUST00000188091.7 ENSMUST00000169382.3 |
Gm21708
Gm21704
|
predicted gene, 21708 predicted gene, 21704 |
| chr10_-_127358231 | 1.52 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr5_-_142594549 | 1.50 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr1_+_15357478 | 1.48 |
ENSMUST00000175681.3
|
Kcnb2
|
potassium voltage gated channel, Shab-related subfamily, member 2 |
| chr2_+_85420854 | 1.45 |
ENSMUST00000052307.5
|
Olfr998
|
olfactory receptor 998 |
| chr4_+_150938376 | 1.44 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr8_+_26091607 | 1.42 |
ENSMUST00000155861.8
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr2_-_163239865 | 1.40 |
ENSMUST00000017961.11
ENSMUST00000109425.3 |
Jph2
|
junctophilin 2 |
| chr9_+_72892786 | 1.38 |
ENSMUST00000156879.8
|
Ccpg1
|
cell cycle progression 1 |
| chr10_-_127358300 | 1.35 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_-_133203838 | 1.33 |
ENSMUST00000033275.4
|
Tex36
|
testis expressed 36 |
| chr16_-_45830575 | 1.32 |
ENSMUST00000130481.2
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr15_+_101013704 | 1.32 |
ENSMUST00000229954.2
|
Ankrd33
|
ankyrin repeat domain 33 |
| chr2_+_71884943 | 1.32 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr7_+_30014235 | 1.30 |
ENSMUST00000054594.15
ENSMUST00000177078.8 ENSMUST00000176504.8 ENSMUST00000176304.8 |
Syne4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr19_-_46661321 | 1.28 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr11_-_69811347 | 1.26 |
ENSMUST00000108610.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_+_157334347 | 1.26 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_157334298 | 1.24 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr11_+_115494751 | 1.24 |
ENSMUST00000058109.9
|
Mrps7
|
mitchondrial ribosomal protein S7 |
| chr14_+_67470884 | 1.24 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr9_+_24194729 | 1.23 |
ENSMUST00000154644.2
|
Npsr1
|
neuropeptide S receptor 1 |
| chr13_-_32960379 | 1.21 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr10_-_95337783 | 1.16 |
ENSMUST00000075829.3
ENSMUST00000217777.2 ENSMUST00000218893.2 |
Mrpl42
|
mitochondrial ribosomal protein L42 |
| chr15_-_77854711 | 1.15 |
ENSMUST00000230419.2
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr15_-_76906150 | 1.15 |
ENSMUST00000230031.2
|
Mb
|
myoglobin |
| chr11_+_58668915 | 1.14 |
ENSMUST00000081533.5
|
Olfr315
|
olfactory receptor 315 |
| chr19_-_46661501 | 1.13 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr11_+_67689094 | 1.12 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr14_-_10185787 | 1.10 |
ENSMUST00000225871.2
|
Gm49355
|
predicted gene, 49355 |
| chr14_+_67470735 | 1.10 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr12_+_10419967 | 1.09 |
ENSMUST00000143739.9
ENSMUST00000002456.10 ENSMUST00000219826.2 ENSMUST00000217944.2 ENSMUST00000218339.3 ENSMUST00000118657.8 ENSMUST00000223534.2 |
Nt5c1b
|
5'-nucleotidase, cytosolic IB |
| chr6_-_122317156 | 1.09 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr2_+_127429125 | 1.09 |
ENSMUST00000028852.13
|
Mrps5
|
mitochondrial ribosomal protein S5 |
| chr7_+_46636562 | 1.08 |
ENSMUST00000185832.2
|
Gm9999
|
predicted gene 9999 |
| chr13_-_21823691 | 1.05 |
ENSMUST00000043081.3
|
Olfr11
|
olfactory receptor 11 |
| chr2_+_65760477 | 1.03 |
ENSMUST00000176109.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr13_+_8935974 | 0.98 |
ENSMUST00000177397.8
ENSMUST00000177400.8 ENSMUST00000177447.2 |
Idi1
|
isopentenyl-diphosphate delta isomerase |
| chr14_-_20714634 | 0.98 |
ENSMUST00000119483.2
|
Synpo2l
|
synaptopodin 2-like |
| chr2_+_85597442 | 0.95 |
ENSMUST00000216397.3
|
Olfr1013
|
olfactory receptor 1013 |
| chr10_+_127595590 | 0.94 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr16_-_58940431 | 0.92 |
ENSMUST00000072608.2
|
Olfr194
|
olfactory receptor 194 |
| chr5_-_66776095 | 0.92 |
ENSMUST00000162366.8
ENSMUST00000162994.8 ENSMUST00000159512.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr13_+_81034214 | 0.91 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr4_+_85972125 | 0.91 |
ENSMUST00000107178.9
ENSMUST00000048885.12 ENSMUST00000141889.8 ENSMUST00000120678.2 |
Adamtsl1
|
ADAMTS-like 1 |
| chr10_+_110756031 | 0.90 |
ENSMUST00000220409.2
ENSMUST00000219502.2 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr17_-_48739874 | 0.89 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr17_-_37404764 | 0.89 |
ENSMUST00000087144.5
|
Olfr91
|
olfactory receptor 91 |
| chr10_+_97442727 | 0.89 |
ENSMUST00000105286.4
|
Kera
|
keratocan |
| chr4_-_118792037 | 0.87 |
ENSMUST00000081960.5
|
Olfr1328
|
olfactory receptor 1328 |
| chr6_-_148732893 | 0.84 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr14_+_63235512 | 0.83 |
ENSMUST00000100492.5
|
Defb47
|
defensin beta 47 |
| chrX_+_75436956 | 0.83 |
ENSMUST00000101419.2
ENSMUST00000178974.2 |
Cldn34b4
|
claudin 34B4 |
| chr14_+_74969737 | 0.83 |
ENSMUST00000022573.17
ENSMUST00000175712.8 ENSMUST00000176957.8 |
Esd
|
esterase D/formylglutathione hydrolase |
| chr5_-_66775979 | 0.81 |
ENSMUST00000162382.8
ENSMUST00000159786.8 ENSMUST00000162349.8 ENSMUST00000087256.12 ENSMUST00000160103.8 ENSMUST00000160870.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr11_+_109304735 | 0.77 |
ENSMUST00000055404.8
|
9930022D16Rik
|
RIKEN cDNA 9930022D16 gene |
| chr4_-_49681954 | 0.77 |
ENSMUST00000029991.3
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| chrX_+_42680037 | 0.76 |
ENSMUST00000105113.4
|
Tex13c1
|
TEX13 family member C1 |
| chrX_-_140508177 | 0.76 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr2_+_118603247 | 0.75 |
ENSMUST00000061360.4
ENSMUST00000130293.8 |
Phgr1
|
proline/histidine/glycine-rich 1 |
| chr9_-_119654522 | 0.75 |
ENSMUST00000070617.8
|
Scn11a
|
sodium channel, voltage-gated, type XI, alpha |
| chr13_+_8935537 | 0.74 |
ENSMUST00000169314.9
|
Idi1
|
isopentenyl-diphosphate delta isomerase |
| chr15_+_81686622 | 0.73 |
ENSMUST00000109553.10
|
Tef
|
thyrotroph embryonic factor |
| chr11_+_58648430 | 0.73 |
ENSMUST00000203731.2
|
Olfr316
|
olfactory receptor 316 |
| chr10_-_128236317 | 0.72 |
ENSMUST00000167859.2
ENSMUST00000218858.2 |
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr15_-_91075933 | 0.72 |
ENSMUST00000069511.8
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr7_+_142662931 | 0.68 |
ENSMUST00000187213.2
|
Kcnq1
|
potassium voltage-gated channel, subfamily Q, member 1 |
| chr17_+_38231439 | 0.68 |
ENSMUST00000216440.2
|
Olfr128
|
olfactory receptor 128 |
| chr7_+_30157704 | 0.68 |
ENSMUST00000126297.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr1_+_98348817 | 0.68 |
ENSMUST00000027575.13
ENSMUST00000160796.2 |
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr7_-_23206631 | 0.67 |
ENSMUST00000227713.2
|
Vmn1r167
|
vomeronasal 1 receptor 167 |
| chr2_+_59442378 | 0.67 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr2_-_26962187 | 0.66 |
ENSMUST00000009358.9
|
Mymk
|
myomaker, myoblast fusion factor |
| chr19_+_5100475 | 0.66 |
ENSMUST00000225427.2
|
Rin1
|
Ras and Rab interactor 1 |
| chr7_-_103420801 | 0.66 |
ENSMUST00000106878.3
|
Olfr69
|
olfactory receptor 69 |
| chr11_+_43046476 | 0.66 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr15_-_77854988 | 0.66 |
ENSMUST00000100484.6
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr12_+_105157302 | 0.65 |
ENSMUST00000090990.6
|
Tcl1b3
|
T cell leukemia/lymphoma 1B, 3 |
| chr1_-_80439165 | 0.65 |
ENSMUST00000211023.2
|
Gm45261
|
predicted gene 45261 |
| chr7_+_17799889 | 0.64 |
ENSMUST00000108483.2
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr4_-_116024788 | 0.63 |
ENSMUST00000030465.10
ENSMUST00000143426.2 |
Tspan1
|
tetraspanin 1 |
| chr3_+_93733327 | 0.63 |
ENSMUST00000081780.4
|
Tdpoz3
|
TD and POZ domain containing 3 |
| chr2_+_91541197 | 0.62 |
ENSMUST00000128140.2
ENSMUST00000140183.2 |
Harbi1
|
harbinger transposase derived 1 |
| chr4_-_96236887 | 0.60 |
ENSMUST00000015368.8
|
Cyp2j11
|
cytochrome P450, family 2, subfamily j, polypeptide 11 |
| chr2_+_65499097 | 0.60 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr9_+_89093210 | 0.59 |
ENSMUST00000118870.8
ENSMUST00000085256.8 |
Mthfs
|
5, 10-methenyltetrahydrofolate synthetase |
| chr7_+_140190081 | 0.59 |
ENSMUST00000072655.3
|
Olfr46
|
olfactory receptor 46 |
| chr2_-_79959178 | 0.59 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_-_42097503 | 0.59 |
ENSMUST00000032648.5
|
4933421I07Rik
|
RIKEN cDNA 4933421I07 gene |
| chr6_+_116490474 | 0.58 |
ENSMUST00000218028.2
ENSMUST00000220134.2 |
Olfr212
|
olfactory receptor 212 |
| chr9_-_39862065 | 0.57 |
ENSMUST00000054067.4
|
Olfr975
|
olfactory receptor 975 |
| chr3_-_10366229 | 0.56 |
ENSMUST00000119761.2
ENSMUST00000029043.13 |
Fabp12
|
fatty acid binding protein 12 |
| chr16_+_93424913 | 0.55 |
ENSMUST00000120115.3
|
Gm5678
|
predicted gene 5678 |
| chr13_-_101831020 | 0.55 |
ENSMUST00000185795.2
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr11_-_69811890 | 0.54 |
ENSMUST00000108609.8
ENSMUST00000108608.8 ENSMUST00000164359.8 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr3_-_151468529 | 0.53 |
ENSMUST00000046739.6
|
Ifi44l
|
interferon-induced protein 44 like |
| chr2_+_91541245 | 0.53 |
ENSMUST00000142692.2
ENSMUST00000090608.6 |
Harbi1
|
harbinger transposase derived 1 |
| chr10_+_127256192 | 0.52 |
ENSMUST00000171434.8
|
R3hdm2
|
R3H domain containing 2 |
| chr10_+_127685785 | 0.52 |
ENSMUST00000077530.3
|
Rdh19
|
retinol dehydrogenase 19 |
| chr11_+_73244561 | 0.51 |
ENSMUST00000108465.4
|
Olfr20
|
olfactory receptor 20 |
| chr14_-_75830550 | 0.50 |
ENSMUST00000164082.9
ENSMUST00000169658.9 |
Cby2
|
chibby family member 2 |
| chr9_-_88601866 | 0.50 |
ENSMUST00000113110.5
|
Mthfsl
|
5, 10-methenyltetrahydrofolate synthetase-like |
| chr7_-_23510068 | 0.48 |
ENSMUST00000228383.2
|
Vmn1r175
|
vomeronasal 1 receptor 175 |
| chr5_+_114142842 | 0.48 |
ENSMUST00000161610.6
|
Dao
|
D-amino acid oxidase |
| chrX_+_61511597 | 0.48 |
ENSMUST00000033537.2
|
4931400O07Rik
|
RIKEN cDNA 4931400O07 gene |
| chr7_+_17799849 | 0.48 |
ENSMUST00000032520.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr10_+_59715439 | 0.48 |
ENSMUST00000142819.8
ENSMUST00000020309.7 |
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr7_-_44397473 | 0.48 |
ENSMUST00000120074.8
|
Zfp473
|
zinc finger protein 473 |
| chr9_-_39868365 | 0.48 |
ENSMUST00000169307.3
ENSMUST00000217360.2 |
Olfr976
|
olfactory receptor 976 |
| chr14_-_55231998 | 0.48 |
ENSMUST00000227518.2
ENSMUST00000226424.2 ENSMUST00000153783.2 ENSMUST00000102803.11 ENSMUST00000168485.8 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr5_-_5529119 | 0.47 |
ENSMUST00000115447.2
|
Pttg1ip2
|
PTTG1IP family member 2 |
| chr9_+_72892850 | 0.47 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr18_-_35841435 | 0.46 |
ENSMUST00000236738.2
ENSMUST00000237995.2 |
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr9_-_48391838 | 0.45 |
ENSMUST00000216470.2
ENSMUST00000217037.2 ENSMUST00000034524.5 ENSMUST00000213895.2 |
Rexo2
|
RNA exonuclease 2 |
| chr2_+_86655007 | 0.45 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr10_-_128236366 | 0.45 |
ENSMUST00000219131.2
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chrY_+_84109980 | 0.44 |
ENSMUST00000177775.2
|
Gm21095
|
predicted gene, 21095 |
| chr11_-_6588637 | 0.44 |
ENSMUST00000102910.4
|
Wap
|
whey acidic protein |
| chr5_-_25047577 | 0.44 |
ENSMUST00000030787.9
|
Rheb
|
Ras homolog enriched in brain |
| chr2_-_101479846 | 0.44 |
ENSMUST00000078494.6
ENSMUST00000160722.8 ENSMUST00000160037.8 |
Rag1
Iftap
|
recombination activating 1 intraflagellar transport associated protein |
| chrY_+_77705501 | 0.44 |
ENSMUST00000179073.2
|
Gm21650
|
predicted gene, 21650 |
| chr19_-_12219023 | 0.43 |
ENSMUST00000087818.2
|
Olfr262
|
olfactory receptor 262 |
| chrY_-_72256967 | 0.43 |
ENSMUST00000178505.2
|
Gm20843
|
predicted gene, 20843 |
| chrY_-_68304748 | 0.43 |
ENSMUST00000180329.2
|
Gm20937
|
predicted gene, 20937 |
| chr6_+_91881193 | 0.43 |
ENSMUST00000205686.2
|
4930590J08Rik
|
RIKEN cDNA 4930590J08 gene |
| chrY_+_55211732 | 0.42 |
ENSMUST00000180249.2
|
Gm20931
|
predicted gene, 20931 |
| chr3_+_63148887 | 0.42 |
ENSMUST00000194324.6
|
Mme
|
membrane metallo endopeptidase |
| chr17_-_46798566 | 0.42 |
ENSMUST00000047034.9
|
Ttbk1
|
tau tubulin kinase 1 |
| chrY_+_70454574 | 0.42 |
ENSMUST00000178934.2
|
Gm20888
|
predicted gene, 20888 |
| chrY_+_79332266 | 0.41 |
ENSMUST00000178063.2
|
Gm20916
|
predicted gene, 20916 |
| chrY_+_49569158 | 0.41 |
ENSMUST00000178556.2
|
Gm21209
|
predicted gene, 21209 |
| chrY_+_62200817 | 0.41 |
ENSMUST00000178115.2
|
Gm21518
|
predicted gene, 21518 |
| chrY_+_55729767 | 0.41 |
ENSMUST00000177834.2
|
Gm21858
|
predicted gene, 21858 |
| chrX_-_153673263 | 0.41 |
ENSMUST00000096852.5
|
Cldn34b1
|
claudin 34B1 |
| chrY_+_51123234 | 0.41 |
ENSMUST00000180133.2
|
Gm21117
|
predicted gene, 21117 |
| chrY_+_65387652 | 0.40 |
ENSMUST00000178198.2
|
Gm20736
|
predicted gene, 20736 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.5 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 2.0 | 12.2 | GO:0002159 | desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) |
| 1.5 | 10.8 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.0 | 3.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 1.0 | 2.9 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.8 | 2.5 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.6 | 18.9 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.6 | 3.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.5 | 3.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.5 | 1.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.4 | 5.4 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.4 | 3.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.4 | 3.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 1.2 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.3 | 7.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.3 | 0.8 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.3 | 1.3 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.3 | 2.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 2.5 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 5.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 1.4 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.2 | 1.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 3.7 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 35.9 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.2 | 0.7 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.2 | 0.9 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 | 0.7 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.2 | 4.4 | GO:1902932 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) positive regulation of alcohol biosynthetic process(GO:1902932) |
| 0.2 | 1.7 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 3.0 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.2 | 5.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 1.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 0.5 | GO:0009078 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 0.5 | GO:0014728 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 0.1 | 4.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 5.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 3.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 2.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 2.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.4 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 1.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.4 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.7 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 3.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 2.3 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.1 | 0.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.9 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 7.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 1.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 1.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.8 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 1.0 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.4 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 1.8 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 1.3 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.1 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.0 | 1.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 4.4 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 0.3 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.8 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.3 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.9 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.7 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.3 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 2.1 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 0.2 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 2.0 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.3 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.0 | 15.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.0 | 0.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 10.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 1.1 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.3 | 12.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 2.8 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 34.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 2.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 3.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 1.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 3.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 4.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 7.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.7 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 7.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 3.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 7.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 10.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 4.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 11.9 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 1.0 | 2.9 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.8 | 4.6 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.8 | 12.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.6 | 4.4 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.6 | 1.8 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.6 | 2.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.5 | 12.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.5 | 3.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.5 | 2.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.4 | 5.3 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.4 | 1.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.3 | 1.7 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.2 | 10.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 2.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 3.0 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 0.9 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.2 | 3.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 1.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 2.2 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.2 | 4.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.5 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 1.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 1.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.8 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 1.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 7.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 3.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 1.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 2.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.0 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 31.7 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 1.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 1.9 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 6.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 2.2 | GO:0048037 | cofactor binding(GO:0048037) |
| 0.0 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 14.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 3.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 11.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 3.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 4.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 3.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 3.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 5.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |