Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
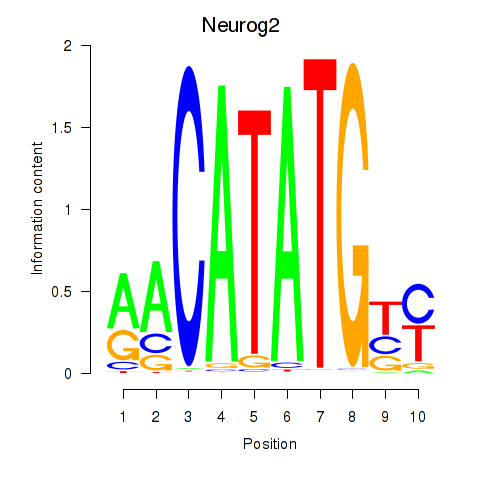
Results for Neurog2
Z-value: 1.36
Transcription factors associated with Neurog2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurog2
|
ENSMUSG00000027967.9 | Neurog2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurog2 | mm39_v1_chr3_+_127426783_127426789 | -0.37 | 1.3e-03 | Click! |
Activity profile of Neurog2 motif
Sorted Z-values of Neurog2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurog2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_118603247 | 27.65 |
ENSMUST00000061360.4
ENSMUST00000130293.8 |
Phgr1
|
proline/histidine/glycine-rich 1 |
| chr8_-_107792264 | 15.97 |
ENSMUST00000034393.7
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr7_+_130633776 | 15.73 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr3_-_123029782 | 13.48 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chr7_+_30399208 | 13.42 |
ENSMUST00000013227.8
|
2200002J24Rik
|
RIKEN cDNA 2200002J24 gene |
| chr6_-_68609426 | 13.41 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr3_+_3573084 | 11.94 |
ENSMUST00000108393.8
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr3_+_90434160 | 11.55 |
ENSMUST00000199538.5
ENSMUST00000164481.7 ENSMUST00000167598.6 |
S100a14
|
S100 calcium binding protein A14 |
| chr12_-_114104740 | 11.49 |
ENSMUST00000103473.2
|
Ighv9-3
|
immunoglobulin heavy variable V9-3 |
| chr13_+_49697919 | 11.08 |
ENSMUST00000177948.2
ENSMUST00000021820.14 |
Aspn
|
asporin |
| chr3_+_122688721 | 10.48 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr9_-_70328816 | 9.74 |
ENSMUST00000034742.8
|
Ccnb2
|
cyclin B2 |
| chr12_-_114878652 | 8.97 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr9_-_21829385 | 8.75 |
ENSMUST00000128442.2
ENSMUST00000119055.8 ENSMUST00000122211.8 ENSMUST00000115351.10 |
Rab3d
|
RAB3D, member RAS oncogene family |
| chr16_+_57173456 | 8.72 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr8_-_107064615 | 8.48 |
ENSMUST00000067512.8
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr17_-_31363245 | 8.48 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr13_+_54849268 | 8.27 |
ENSMUST00000037145.8
|
Cdhr2
|
cadherin-related family member 2 |
| chr2_-_163239865 | 7.90 |
ENSMUST00000017961.11
ENSMUST00000109425.3 |
Jph2
|
junctophilin 2 |
| chr1_+_93301596 | 7.86 |
ENSMUST00000058682.11
ENSMUST00000186641.7 |
Ano7
|
anoctamin 7 |
| chr3_-_123029745 | 7.24 |
ENSMUST00000106426.8
|
Synpo2
|
synaptopodin 2 |
| chr12_-_114901026 | 7.23 |
ENSMUST00000103516.2
ENSMUST00000191868.2 |
Ighv1-42
|
immunoglobulin heavy variable V1-42 |
| chr7_-_4606104 | 7.21 |
ENSMUST00000049113.14
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr11_+_43046476 | 7.13 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr6_-_113696390 | 6.99 |
ENSMUST00000203588.2
ENSMUST00000204163.3 ENSMUST00000203363.3 |
Ghrl
|
ghrelin |
| chrX_+_139808351 | 6.73 |
ENSMUST00000033806.5
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr9_-_99599312 | 6.51 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr12_-_114646685 | 6.27 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr12_-_114710326 | 6.14 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr2_+_24235300 | 5.55 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr8_+_123920682 | 5.52 |
ENSMUST00000212409.2
|
Dpep1
|
dipeptidase 1 |
| chr2_+_57887896 | 5.46 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr7_+_140659038 | 5.35 |
ENSMUST00000159375.8
|
Pkp3
|
plakophilin 3 |
| chr12_-_114815280 | 5.24 |
ENSMUST00000103512.3
|
Ighv1-34
|
immunoglobulin heavy variable 1-34 |
| chr4_-_141325517 | 5.20 |
ENSMUST00000131317.8
ENSMUST00000006381.11 ENSMUST00000129602.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr6_-_83010402 | 5.18 |
ENSMUST00000089651.6
|
Dok1
|
docking protein 1 |
| chr5_+_129173804 | 4.94 |
ENSMUST00000056617.14
ENSMUST00000156437.3 |
Adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr12_-_115706126 | 4.90 |
ENSMUST00000166645.2
ENSMUST00000196690.2 |
Ighv1-71
|
immunoglobulin heavy variable 1-71 |
| chr6_-_52203146 | 4.64 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr12_-_115276219 | 4.64 |
ENSMUST00000103529.4
|
Ighv1-58
|
immunoglobulin heavy variable 1-58 |
| chr6_+_68414401 | 4.61 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr6_+_41331039 | 4.28 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr11_-_69696428 | 4.28 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr10_-_128237087 | 4.20 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr17_+_36176485 | 3.81 |
ENSMUST00000127442.8
ENSMUST00000144382.8 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr5_+_90638580 | 3.76 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chrX_+_55500170 | 3.74 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr7_+_4928784 | 3.70 |
ENSMUST00000057612.9
|
Ssc5d
|
scavenger receptor cysteine rich family, 5 domains |
| chr2_+_125876566 | 3.64 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr11_+_115225557 | 3.50 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr11_-_72441054 | 3.49 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr10_+_110756031 | 3.40 |
ENSMUST00000220409.2
ENSMUST00000219502.2 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr12_-_114073050 | 3.34 |
ENSMUST00000103472.4
|
Ighv9-2
|
immunoglobulin heavy variable V9-2 |
| chrX_-_7054952 | 3.29 |
ENSMUST00000004428.14
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr3_-_10366229 | 3.24 |
ENSMUST00000119761.2
ENSMUST00000029043.13 |
Fabp12
|
fatty acid binding protein 12 |
| chr17_-_48739874 | 3.19 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr12_-_115410489 | 3.19 |
ENSMUST00000194581.2
|
Ighv1-62-2
|
immunoglobulin heavy variable 1-62-2 |
| chr11_-_69811717 | 3.15 |
ENSMUST00000152589.2
ENSMUST00000108612.8 ENSMUST00000108611.8 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr18_-_35781422 | 3.07 |
ENSMUST00000237462.2
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr11_-_97673203 | 3.06 |
ENSMUST00000128801.2
ENSMUST00000103146.5 |
Rpl23
|
ribosomal protein L23 |
| chr9_+_111011327 | 3.03 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr12_-_114057841 | 3.00 |
ENSMUST00000103471.2
ENSMUST00000195884.2 |
Ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr16_+_96006919 | 2.97 |
ENSMUST00000129904.3
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein |
| chr1_-_74343543 | 2.97 |
ENSMUST00000016309.16
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr3_-_95902949 | 2.96 |
ENSMUST00000123006.8
ENSMUST00000130043.8 |
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr15_-_89294434 | 2.94 |
ENSMUST00000109314.9
|
Syce3
|
synaptonemal complex central element protein 3 |
| chr9_-_58066484 | 2.94 |
ENSMUST00000041477.15
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr17_+_35345292 | 2.91 |
ENSMUST00000061859.7
|
D17H6S53E
|
DNA segment, Chr 17, human D6S53E |
| chr5_-_5529119 | 2.88 |
ENSMUST00000115447.2
|
Pttg1ip2
|
PTTG1IP family member 2 |
| chr10_-_128236317 | 2.83 |
ENSMUST00000167859.2
ENSMUST00000218858.2 |
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr19_-_53360197 | 2.82 |
ENSMUST00000086887.2
|
Gm10197
|
predicted gene 10197 |
| chr1_-_74343471 | 2.69 |
ENSMUST00000113796.8
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr4_+_11579648 | 2.69 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr3_-_92481033 | 2.68 |
ENSMUST00000053107.6
|
Ivl
|
involucrin |
| chr10_-_128236366 | 2.66 |
ENSMUST00000219131.2
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr9_+_66853343 | 2.61 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr11_-_97886997 | 2.51 |
ENSMUST00000042971.16
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr4_-_144190326 | 2.49 |
ENSMUST00000105749.2
|
Aadacl3
|
arylacetamide deacetylase like 3 |
| chr17_-_35293447 | 2.49 |
ENSMUST00000007259.4
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr9_-_58065800 | 2.48 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr6_-_128252540 | 2.48 |
ENSMUST00000130454.8
|
Tead4
|
TEA domain family member 4 |
| chr4_-_49681954 | 2.46 |
ENSMUST00000029991.3
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| chr14_-_70405288 | 2.33 |
ENSMUST00000129174.8
ENSMUST00000125300.3 |
Pdlim2
|
PDZ and LIM domain 2 |
| chr2_+_74557418 | 2.33 |
ENSMUST00000111980.4
|
Hoxd4
|
homeobox D4 |
| chr7_+_127503812 | 2.32 |
ENSMUST00000151451.3
ENSMUST00000124533.3 ENSMUST00000206745.2 ENSMUST00000206140.2 |
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr7_+_43361930 | 2.29 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr3_-_63806794 | 2.28 |
ENSMUST00000161052.3
ENSMUST00000159188.2 ENSMUST00000177143.8 |
Plch1
|
phospholipase C, eta 1 |
| chr7_-_126736979 | 2.24 |
ENSMUST00000049931.6
|
Spn
|
sialophorin |
| chr1_-_170133901 | 2.24 |
ENSMUST00000179801.3
|
Gm7694
|
predicted gene 7694 |
| chr9_+_111011388 | 2.22 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr13_-_67547858 | 2.21 |
ENSMUST00000224684.2
ENSMUST00000181071.8 ENSMUST00000109732.3 ENSMUST00000224825.2 |
Zfp429
|
zinc finger protein 429 |
| chr5_+_3393893 | 2.15 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr19_-_53577499 | 2.14 |
ENSMUST00000095978.5
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chrX_-_153911405 | 2.12 |
ENSMUST00000076671.4
|
Cldn34b2
|
claudin 34B2 |
| chr17_+_29042640 | 2.11 |
ENSMUST00000233088.2
ENSMUST00000233182.2 ENSMUST00000233520.2 |
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr4_-_118314707 | 2.11 |
ENSMUST00000102671.10
|
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr1_-_74343279 | 2.08 |
ENSMUST00000141560.2
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr1_+_132996237 | 2.07 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chrX_+_140258381 | 2.06 |
ENSMUST00000112931.8
ENSMUST00000112930.8 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr4_+_85972125 | 2.06 |
ENSMUST00000107178.9
ENSMUST00000048885.12 ENSMUST00000141889.8 ENSMUST00000120678.2 |
Adamtsl1
|
ADAMTS-like 1 |
| chr6_+_83011154 | 2.05 |
ENSMUST00000000707.9
ENSMUST00000101257.4 |
Loxl3
|
lysyl oxidase-like 3 |
| chr1_+_180762587 | 2.01 |
ENSMUST00000037361.9
|
Lefty1
|
left right determination factor 1 |
| chr9_-_71803354 | 1.99 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr7_-_126303947 | 1.95 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr16_-_75706161 | 1.95 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr17_+_29042544 | 1.93 |
ENSMUST00000140587.9
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr17_-_24662055 | 1.92 |
ENSMUST00000119932.8
ENSMUST00000088506.12 |
Dnase1l2
|
deoxyribonuclease 1-like 2 |
| chr2_-_163486998 | 1.80 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr4_-_82768958 | 1.80 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr2_+_69500444 | 1.78 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr17_+_87224776 | 1.78 |
ENSMUST00000042172.7
|
Tmem247
|
transmembrane protein 247 |
| chr4_-_118314647 | 1.75 |
ENSMUST00000106375.2
ENSMUST00000168404.9 ENSMUST00000006556.11 |
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr11_-_101316156 | 1.73 |
ENSMUST00000103102.10
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr2_-_79287095 | 1.72 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr3_-_107240989 | 1.69 |
ENSMUST00000061772.11
|
Rbm15
|
RNA binding motif protein 15 |
| chr13_-_67523832 | 1.68 |
ENSMUST00000225787.2
ENSMUST00000172266.8 ENSMUST00000057070.9 |
Zfp456
|
zinc finger protein 456 |
| chr2_-_93787441 | 1.65 |
ENSMUST00000099689.5
|
Gm13889
|
predicted gene 13889 |
| chr1_-_164763091 | 1.63 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr16_+_57173632 | 1.61 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr9_+_56344700 | 1.60 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr18_+_60907698 | 1.58 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chr18_+_60907668 | 1.57 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr6_-_40906665 | 1.56 |
ENSMUST00000136499.2
|
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
| chr8_+_95393228 | 1.54 |
ENSMUST00000034228.16
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr9_+_108437485 | 1.53 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr3_-_105940130 | 1.53 |
ENSMUST00000200146.2
|
Chil5
|
chitinase-like 5 |
| chr6_-_85879510 | 1.48 |
ENSMUST00000159755.8
|
Nat8f4
|
N-acetyltransferase 8 (GCN5-related) family member 4 |
| chr7_-_28649094 | 1.47 |
ENSMUST00000148196.3
|
Actn4
|
actinin alpha 4 |
| chr7_-_44397730 | 1.42 |
ENSMUST00000118162.8
ENSMUST00000140599.9 ENSMUST00000120798.8 |
Zfp473
|
zinc finger protein 473 |
| chr6_-_112364974 | 1.41 |
ENSMUST00000238755.2
ENSMUST00000060847.6 |
Ssu2
|
ssu-2 homolog (C. elegans) |
| chr15_-_85695810 | 1.40 |
ENSMUST00000144067.8
|
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr1_+_170104889 | 1.37 |
ENSMUST00000179976.3
|
Sh2d1b1
|
SH2 domain containing 1B1 |
| chr11_-_69811347 | 1.36 |
ENSMUST00000108610.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr17_-_65946817 | 1.34 |
ENSMUST00000233702.2
|
Txndc2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr18_+_42644552 | 1.32 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr11_+_117157024 | 1.30 |
ENSMUST00000019038.15
|
Septin9
|
septin 9 |
| chr18_+_61044830 | 1.28 |
ENSMUST00000040359.6
|
Arsi
|
arylsulfatase i |
| chr2_+_75662511 | 1.20 |
ENSMUST00000047232.14
ENSMUST00000111952.9 |
Agps
|
alkylglycerone phosphate synthase |
| chr14_-_42872221 | 1.18 |
ENSMUST00000100694.5
|
Gm10376
|
predicted gene 10376 |
| chr3_+_105821450 | 1.18 |
ENSMUST00000198080.5
ENSMUST00000199977.2 |
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr12_+_86781154 | 1.18 |
ENSMUST00000095527.6
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chrX_+_35592006 | 1.18 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr15_-_85695855 | 1.18 |
ENSMUST00000134631.8
ENSMUST00000154814.2 ENSMUST00000071876.13 ENSMUST00000150995.8 |
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr11_+_53324126 | 1.17 |
ENSMUST00000018382.7
|
Gdf9
|
growth differentiation factor 9 |
| chr12_-_114909863 | 1.13 |
ENSMUST00000103517.2
ENSMUST00000195417.2 |
Ighv1-43
|
immunoglobulin heavy variable V1-43 |
| chr9_-_119151428 | 1.10 |
ENSMUST00000040853.11
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr7_-_44397473 | 1.03 |
ENSMUST00000120074.8
|
Zfp473
|
zinc finger protein 473 |
| chr1_+_187995096 | 1.01 |
ENSMUST00000060479.14
|
Ush2a
|
usherin |
| chr3_+_84081411 | 1.00 |
ENSMUST00000193882.2
|
Gm6525
|
predicted pseudogene 6525 |
| chr2_+_163535925 | 1.00 |
ENSMUST00000109400.3
|
Pkig
|
protein kinase inhibitor, gamma |
| chr2_-_110983281 | 0.99 |
ENSMUST00000132464.2
|
Gm15130
|
predicted gene 15130 |
| chr2_-_93787383 | 0.97 |
ENSMUST00000148314.3
|
Gm13889
|
predicted gene 13889 |
| chr1_-_161807205 | 0.94 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr7_+_140190081 | 0.92 |
ENSMUST00000072655.3
|
Olfr46
|
olfactory receptor 46 |
| chr16_+_10652910 | 0.91 |
ENSMUST00000037913.9
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr7_-_133203838 | 0.91 |
ENSMUST00000033275.4
|
Tex36
|
testis expressed 36 |
| chr17_+_18108086 | 0.91 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr14_+_47535717 | 0.90 |
ENSMUST00000166743.9
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr12_+_86781141 | 0.89 |
ENSMUST00000223308.2
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr11_+_31822211 | 0.85 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_+_25293140 | 0.84 |
ENSMUST00000154809.8
ENSMUST00000055921.14 ENSMUST00000141567.8 |
Npdc1
|
neural proliferation, differentiation and control 1 |
| chr11_+_120123727 | 0.84 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr7_+_127187910 | 0.82 |
ENSMUST00000205694.2
ENSMUST00000033088.8 ENSMUST00000206914.2 |
Rnf40
|
ring finger protein 40 |
| chr4_-_52936281 | 0.80 |
ENSMUST00000217546.2
|
Olfr271-ps1
|
olfactory receptor 271, pseudogene 1 |
| chr15_+_81912312 | 0.79 |
ENSMUST00000230729.2
|
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr14_+_31750946 | 0.79 |
ENSMUST00000022460.11
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chr16_-_26810402 | 0.78 |
ENSMUST00000231299.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr7_-_85971258 | 0.75 |
ENSMUST00000044256.6
|
Olfr308
|
olfactory receptor 308 |
| chr9_+_106158212 | 0.74 |
ENSMUST00000072206.14
|
Poc1a
|
POC1 centriolar protein A |
| chrX_-_52610946 | 0.74 |
ENSMUST00000123034.3
|
4933416I08Rik
|
RIKEN cDNA 4933416I08 gene |
| chr7_+_104228831 | 0.73 |
ENSMUST00000098174.3
|
Olfr653
|
olfactory receptor 653 |
| chr2_-_180284468 | 0.73 |
ENSMUST00000037877.11
|
Tcfl5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr2_+_85519775 | 0.72 |
ENSMUST00000054868.2
|
Olfr1008
|
olfactory receptor 1008 |
| chr2_-_38955452 | 0.71 |
ENSMUST00000112850.9
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr8_+_11890474 | 0.69 |
ENSMUST00000033909.14
ENSMUST00000209692.2 |
Tex29
|
testis expressed 29 |
| chrX_-_72703652 | 0.69 |
ENSMUST00000114472.8
|
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr8_+_95393349 | 0.68 |
ENSMUST00000109527.6
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chrX_+_153264998 | 0.68 |
ENSMUST00000026029.2
|
Samt4
|
spermatogenesis associated multipass transmembrane protein 4 |
| chr11_+_95734028 | 0.68 |
ENSMUST00000107709.8
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr2_-_76700830 | 0.67 |
ENSMUST00000138542.2
|
Ttn
|
titin |
| chr6_+_112250719 | 0.65 |
ENSMUST00000032376.6
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr6_+_68657317 | 0.64 |
ENSMUST00000198735.2
|
Igkv10-95
|
immunoglobulin kappa variable 10-95 |
| chrY_-_3410148 | 0.64 |
ENSMUST00000190283.7
ENSMUST00000188091.7 ENSMUST00000169382.3 |
Gm21708
Gm21704
|
predicted gene, 21708 predicted gene, 21704 |
| chr10_-_116385007 | 0.63 |
ENSMUST00000164088.8
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr11_+_58668915 | 0.62 |
ENSMUST00000081533.5
|
Olfr315
|
olfactory receptor 315 |
| chr12_+_108376884 | 0.61 |
ENSMUST00000109857.8
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr12_-_84923252 | 0.60 |
ENSMUST00000163189.8
ENSMUST00000110254.9 ENSMUST00000002073.13 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr10_-_25076008 | 0.59 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr10_+_97442727 | 0.59 |
ENSMUST00000105286.4
|
Kera
|
keratocan |
| chr12_+_108376801 | 0.59 |
ENSMUST00000054955.14
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr5_+_146450933 | 0.58 |
ENSMUST00000200228.5
ENSMUST00000036715.16 ENSMUST00000077133.7 |
Gm3402
|
predicted gene 3402 |
| chr14_-_10185523 | 0.58 |
ENSMUST00000223702.2
ENSMUST00000223762.2 ENSMUST00000112669.10 ENSMUST00000163392.3 |
3830406C13Rik
|
RIKEN cDNA 3830406C13 gene |
| chrY_+_60251163 | 0.57 |
ENSMUST00000178751.2
|
Gm28891
|
predicted gene 28891 |
| chrY_+_47457003 | 0.57 |
ENSMUST00000179672.2
|
Gm21256
|
predicted gene, 21256 |
| chr17_+_37977879 | 0.56 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chrY_-_32574356 | 0.56 |
ENSMUST00000177842.2
|
Gm21094
|
predicted gene, 21094 |
| chrY_+_2830680 | 0.55 |
ENSMUST00000171534.8
ENSMUST00000100360.5 ENSMUST00000179404.8 |
Rbmy
Gm10256
|
RNA binding motif protein, Y chromosome predicted gene 10256 |
| chr2_-_38955518 | 0.55 |
ENSMUST00000039165.15
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr13_+_21364330 | 0.55 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr7_+_44397837 | 0.54 |
ENSMUST00000002275.15
|
Vrk3
|
vaccinia related kinase 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 11.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 1.9 | 9.7 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.9 | 7.7 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 1.7 | 8.7 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 1.7 | 7.0 | GO:2000506 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 1.7 | 8.3 | GO:1904970 | brush border assembly(GO:1904970) |
| 1.5 | 20.7 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.4 | 8.5 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 1.4 | 8.5 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 1.4 | 5.5 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 1.3 | 7.9 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 1.3 | 3.9 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 1.2 | 3.6 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.1 | 5.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.0 | 5.2 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 1.0 | 7.9 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.9 | 6.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.8 | 3.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.8 | 4.5 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.7 | 2.2 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.7 | 3.7 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.7 | 2.2 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.7 | 2.0 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.6 | 12.9 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.6 | 3.1 | GO:2001274 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.6 | 3.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.5 | 1.6 | GO:2000412 | immunoglobulin production in mucosal tissue(GO:0002426) positive regulation of thymocyte migration(GO:2000412) positive regulation of natural killer cell chemotaxis(GO:2000503) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.5 | 2.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.5 | 13.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.5 | 8.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.5 | 2.5 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.4 | 3.5 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.4 | 60.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.4 | 2.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.4 | 4.3 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.4 | 4.9 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.4 | 1.5 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.4 | 1.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.4 | 1.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 1.4 | GO:0002767 | immune response-inhibiting signal transduction(GO:0002765) immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.3 | 5.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.3 | 10.5 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.2 | 3.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 3.8 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 1.2 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.2 | 2.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 5.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 4.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 2.5 | GO:0007320 | insemination(GO:0007320) |
| 0.2 | 1.5 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 3.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 0.8 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 | 1.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.8 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.7 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 1.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.6 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.8 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 0.7 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 2.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.2 | GO:0002654 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.3 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 1.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 5.5 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 2.5 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 1.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 0.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.6 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 1.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 4.3 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.9 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 1.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 4.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 3.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.4 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.5 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 5.6 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.9 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 1.2 | GO:0007052 | mitotic spindle organization(GO:0007052) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 15.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 1.1 | 5.4 | GO:0005914 | spot adherens junction(GO:0005914) |
| 1.0 | 10.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.8 | 8.3 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.7 | 7.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.7 | 2.2 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.6 | 8.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.5 | 60.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 4.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 3.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.4 | 2.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 4.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 2.1 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 1.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 2.9 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 8.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 3.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 30.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.8 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 1.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 5.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 4.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 7.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 2.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 5.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 10.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 21.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 6.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 12.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 10.7 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 2.3 | 7.0 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 1.4 | 5.6 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.2 | 10.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.9 | 3.8 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.9 | 26.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.9 | 3.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.8 | 8.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.8 | 5.5 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.6 | 8.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.6 | 7.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.5 | 2.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.5 | 3.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.5 | 2.0 | GO:0038100 | nodal binding(GO:0038100) |
| 0.5 | 60.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 1.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.4 | 3.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 7.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 3.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 2.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 8.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 6.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 13.2 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.3 | 3.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 2.5 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.3 | 7.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 1.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 4.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 7.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 2.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 9.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 3.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 3.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 6.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 1.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.8 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 11.9 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.1 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 3.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 9.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.6 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 3.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 1.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 2.9 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 1.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 3.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.5 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 2.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 3.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 1.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 2.5 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 3.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.1 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.3 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.8 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 9.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 5.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 5.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 16.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 14.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 13.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 9.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 7.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 5.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 9.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 4.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 6.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.9 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 1.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 5.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 5.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.9 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 2.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 6.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 2.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 2.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |