Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
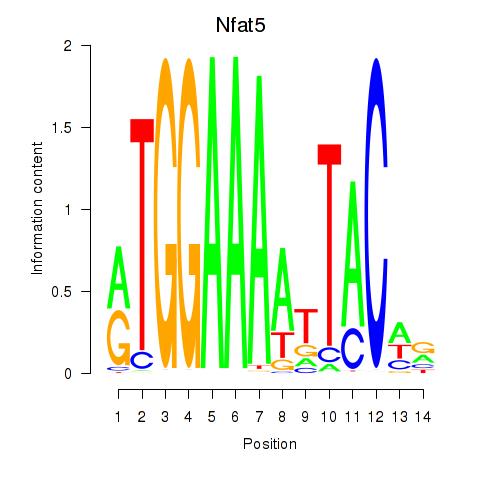
Results for Nfat5
Z-value: 0.99
Transcription factors associated with Nfat5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfat5
|
ENSMUSG00000003847.17 | Nfat5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfat5 | mm39_v1_chr8_+_108020132_108020191 | -0.33 | 4.7e-03 | Click! |
Activity profile of Nfat5 motif
Sorted Z-values of Nfat5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfat5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_129548954 | 9.09 |
ENSMUST00000029653.7
|
Egf
|
epidermal growth factor |
| chr12_+_36042899 | 6.65 |
ENSMUST00000020898.12
|
Agr2
|
anterior gradient 2 |
| chr11_-_99383938 | 6.27 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr8_-_108129829 | 5.70 |
ENSMUST00000003947.9
|
Nqo1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr15_+_99476935 | 5.54 |
ENSMUST00000023752.6
|
Aqp2
|
aquaporin 2 |
| chr1_+_87983099 | 5.19 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_+_83741657 | 4.89 |
ENSMUST00000021016.10
|
Hnf1b
|
HNF1 homeobox B |
| chr11_+_83741689 | 4.50 |
ENSMUST00000108114.9
|
Hnf1b
|
HNF1 homeobox B |
| chr3_-_98631847 | 4.50 |
ENSMUST00000107021.8
ENSMUST00000107022.8 |
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr7_-_80052491 | 4.42 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr17_-_34694911 | 4.31 |
ENSMUST00000065841.5
|
Btnl4
|
butyrophilin-like 4 |
| chr1_+_88334678 | 4.25 |
ENSMUST00000027518.12
|
Spp2
|
secreted phosphoprotein 2 |
| chr17_-_34736326 | 4.07 |
ENSMUST00000075483.5
|
Btnl6
|
butyrophilin-like 6 |
| chr1_+_87983189 | 3.86 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr19_+_25214322 | 3.74 |
ENSMUST00000049400.15
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr19_-_7688628 | 3.73 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr15_-_75766321 | 3.51 |
ENSMUST00000023237.8
|
Naprt
|
nicotinate phosphoribosyltransferase |
| chr11_+_120382666 | 3.23 |
ENSMUST00000026899.4
|
Slc25a10
|
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
| chr7_+_140658101 | 3.19 |
ENSMUST00000106039.9
|
Pkp3
|
plakophilin 3 |
| chr10_+_21870565 | 3.16 |
ENSMUST00000020145.12
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr15_-_57128522 | 2.98 |
ENSMUST00000137764.2
ENSMUST00000022995.13 |
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr15_+_25933632 | 2.87 |
ENSMUST00000228327.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr6_+_48849804 | 2.69 |
ENSMUST00000204856.2
|
Aoc1
|
amine oxidase, copper-containing 1 |
| chr4_-_82423985 | 2.68 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr11_-_88754543 | 2.63 |
ENSMUST00000107904.3
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr18_-_78640066 | 2.62 |
ENSMUST00000235389.2
ENSMUST00000237674.2 |
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr10_+_21869776 | 2.61 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr2_-_30305472 | 2.51 |
ENSMUST00000134120.2
ENSMUST00000102854.10 |
Crat
|
carnitine acetyltransferase |
| chr7_+_19753097 | 2.45 |
ENSMUST00000117909.2
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr14_-_4808744 | 2.26 |
ENSMUST00000022303.17
ENSMUST00000091471.12 |
Thrb
|
thyroid hormone receptor beta |
| chr6_+_91661034 | 2.26 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr5_-_104169696 | 2.25 |
ENSMUST00000119025.2
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr9_-_36549886 | 2.21 |
ENSMUST00000093868.9
ENSMUST00000184611.2 |
Pate14
|
prostate and testis expressed 14 |
| chr3_-_27764571 | 2.20 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr10_+_5589210 | 2.20 |
ENSMUST00000019906.6
|
Vip
|
vasoactive intestinal polypeptide |
| chr1_-_93373145 | 2.15 |
ENSMUST00000186787.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr10_-_95251327 | 2.15 |
ENSMUST00000172070.8
ENSMUST00000150432.8 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr11_+_94172657 | 2.08 |
ENSMUST00000208510.3
|
Gm21885
|
predicted gene, 21885 |
| chr6_-_101354858 | 2.07 |
ENSMUST00000075994.11
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr5_-_104169785 | 2.05 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chrX_+_138585728 | 2.03 |
ENSMUST00000096313.4
|
Tbc1d8b
|
TBC1 domain family, member 8B |
| chr3_+_91921890 | 2.02 |
ENSMUST00000047660.5
|
Pglyrp3
|
peptidoglycan recognition protein 3 |
| chr6_+_91661074 | 2.01 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr11_+_9141934 | 1.99 |
ENSMUST00000042740.13
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr10_-_95251145 | 1.97 |
ENSMUST00000119917.2
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr11_+_118913788 | 1.94 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr7_+_19725390 | 1.90 |
ENSMUST00000207805.2
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr8_-_106706035 | 1.90 |
ENSMUST00000034371.9
|
Dpep3
|
dipeptidase 3 |
| chr2_-_30305313 | 1.86 |
ENSMUST00000132981.9
ENSMUST00000129494.2 |
Crat
|
carnitine acetyltransferase |
| chr7_+_46496929 | 1.85 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr6_+_92793440 | 1.84 |
ENSMUST00000057977.4
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr6_+_73225616 | 1.83 |
ENSMUST00000203632.2
|
Suclg1
|
succinate-CoA ligase, GDP-forming, alpha subunit |
| chr3_-_27764522 | 1.80 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr10_-_29411857 | 1.76 |
ENSMUST00000092623.5
|
Rspo3
|
R-spondin 3 |
| chr2_+_172391501 | 1.74 |
ENSMUST00000030391.9
|
Tfap2c
|
transcription factor AP-2, gamma |
| chr10_-_78188046 | 1.71 |
ENSMUST00000146899.2
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr7_+_19741948 | 1.70 |
ENSMUST00000073151.13
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr16_-_16418397 | 1.69 |
ENSMUST00000159542.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr18_+_61408073 | 1.62 |
ENSMUST00000135688.2
|
Pde6a
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr7_-_126224848 | 1.59 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr5_+_139408906 | 1.56 |
ENSMUST00000066211.5
|
Gper1
|
G protein-coupled estrogen receptor 1 |
| chr14_+_5894220 | 1.41 |
ENSMUST00000063750.8
|
Rarb
|
retinoic acid receptor, beta |
| chr6_+_139564196 | 1.40 |
ENSMUST00000188066.2
ENSMUST00000190962.7 |
Pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr4_-_135300934 | 1.36 |
ENSMUST00000105855.2
|
Grhl3
|
grainyhead like transcription factor 3 |
| chr10_-_125164399 | 1.34 |
ENSMUST00000063318.10
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr4_-_49506538 | 1.33 |
ENSMUST00000043056.9
|
Baat
|
bile acid-Coenzyme A: amino acid N-acyltransferase |
| chr3_-_144804784 | 1.29 |
ENSMUST00000040465.11
ENSMUST00000198993.2 |
Clca2
|
chloride channel accessory 2 |
| chr13_+_19362068 | 1.29 |
ENSMUST00000103553.3
|
Trgv7
|
T cell receptor gamma, variable 7 |
| chr11_+_95152355 | 1.26 |
ENSMUST00000021242.5
|
Tac4
|
tachykinin 4 |
| chr1_+_24717793 | 1.26 |
ENSMUST00000186190.2
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr19_+_56414114 | 1.24 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chrX_+_72030945 | 1.22 |
ENSMUST00000164800.8
ENSMUST00000114546.9 |
Zfp185
|
zinc finger protein 185 |
| chr14_+_63284438 | 1.16 |
ENSMUST00000067990.8
ENSMUST00000111203.2 |
Defb42
|
defensin beta 42 |
| chr11_-_118292678 | 1.16 |
ENSMUST00000106290.4
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr11_-_118292758 | 1.12 |
ENSMUST00000043722.10
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr1_+_165312738 | 1.11 |
ENSMUST00000111440.8
ENSMUST00000027852.15 ENSMUST00000111439.8 |
Adcy10
|
adenylate cyclase 10 |
| chr7_+_46496552 | 1.11 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr18_+_36693646 | 1.06 |
ENSMUST00000155329.9
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr1_+_180773462 | 1.05 |
ENSMUST00000160508.2
ENSMUST00000160536.2 |
Tmem63a
|
transmembrane protein 63a |
| chr4_-_82423944 | 1.04 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr5_-_15083012 | 1.04 |
ENSMUST00000167908.2
|
Gm17019
|
predicted gene 17019 |
| chr5_-_26263579 | 0.98 |
ENSMUST00000095004.4
|
Gm7347
|
predicted gene 7347 |
| chr5_+_17578178 | 0.92 |
ENSMUST00000166086.3
|
Speer4f2
|
spermatogenesis associated glutamate (E)-rich protein 4f2 |
| chr5_-_26147379 | 0.89 |
ENSMUST00000196214.2
|
Gm21663
|
predicted gene, 21663 |
| chr11_+_54261452 | 0.88 |
ENSMUST00000094193.3
|
Meikin
|
meiotic kinetochore factor |
| chrX_-_16683578 | 0.88 |
ENSMUST00000040820.13
|
Maob
|
monoamine oxidase B |
| chr5_-_26326419 | 0.88 |
ENSMUST00000088236.5
|
Gm10220
|
predicted gene 10220 |
| chr10_-_78187887 | 0.86 |
ENSMUST00000105388.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr5_-_26310360 | 0.85 |
ENSMUST00000063524.3
|
5031410I06Rik
|
RIKEN cDNA 5031410I06 gene |
| chr5_-_15028949 | 0.84 |
ENSMUST00000096953.5
|
Gm10354
|
predicted gene 10354 |
| chr8_-_129858142 | 0.83 |
ENSMUST00000108743.2
|
Gm10999
|
predicted gene 10999 |
| chr9_+_3025417 | 0.82 |
ENSMUST00000075573.7
|
Gm10717
|
predicted gene 10717 |
| chr5_-_26227914 | 0.80 |
ENSMUST00000072286.7
|
Gm5862
|
predicted gene 5862 |
| chr12_-_83643964 | 0.79 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr2_+_36886739 | 0.79 |
ENSMUST00000069578.5
|
Olfr357
|
olfactory receptor 357 |
| chr5_-_14964913 | 0.75 |
ENSMUST00000035980.9
|
Gm9758
|
predicted gene 9758 |
| chr7_-_30298287 | 0.74 |
ENSMUST00000108150.2
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr15_+_8997480 | 0.73 |
ENSMUST00000227191.3
|
Ranbp3l
|
RAN binding protein 3-like |
| chr5_-_26177661 | 0.70 |
ENSMUST00000200447.2
|
Gm21680
|
predicted gene, 21680 |
| chr5_-_26193648 | 0.70 |
ENSMUST00000191203.7
|
Gm21698
|
predicted gene, 21698 |
| chr13_-_21327251 | 0.70 |
ENSMUST00000055298.6
|
Olfr1368
|
olfactory receptor 1368 |
| chr5_-_26244556 | 0.68 |
ENSMUST00000079447.4
|
Speer4a
|
spermatogenesis associated glutamate (E)-rich protein 4A |
| chr8_-_76133212 | 0.66 |
ENSMUST00000212864.2
|
Gm10358
|
predicted gene 10358 |
| chr4_+_19575128 | 0.66 |
ENSMUST00000108253.8
ENSMUST00000029888.4 |
Rmdn1
|
regulator of microtubule dynamics 1 |
| chr18_+_57666852 | 0.66 |
ENSMUST00000079738.10
ENSMUST00000135806.8 ENSMUST00000127130.9 |
Ccdc192
|
coiled-coil domain containing 192 |
| chr13_+_27241551 | 0.65 |
ENSMUST00000110369.10
ENSMUST00000224228.2 ENSMUST00000018061.7 |
Prl
|
prolactin |
| chr19_+_34078333 | 0.64 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr4_+_3940747 | 0.62 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr12_-_83643883 | 0.61 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr5_-_26159415 | 0.60 |
ENSMUST00000162387.6
|
Gm21671
|
predicted gene, 21671 |
| chrX_+_37861548 | 0.60 |
ENSMUST00000050744.6
|
6030498E09Rik
|
RIKEN cDNA 6030498E09 gene |
| chr5_-_27706360 | 0.59 |
ENSMUST00000155721.2
ENSMUST00000053257.10 |
Speer4b
|
spermatogenesis associated glutamate (E)-rich protein 4B |
| chr2_+_176522933 | 0.59 |
ENSMUST00000126726.2
ENSMUST00000122218.9 |
Gm14408
|
predicted gene 14408 |
| chr7_+_46496506 | 0.58 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr13_-_120374288 | 0.57 |
ENSMUST00000179502.2
|
Gm21761
|
predicted gene, 21761 |
| chr2_+_32127309 | 0.57 |
ENSMUST00000123740.3
|
Pomt1
|
protein-O-mannosyltransferase 1 |
| chr15_-_38079089 | 0.56 |
ENSMUST00000110336.4
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chrX_-_93408176 | 0.55 |
ENSMUST00000239046.2
|
CXorf58
|
family with sequence similarity 90, member A1B |
| chr5_-_66775979 | 0.55 |
ENSMUST00000162382.8
ENSMUST00000159786.8 ENSMUST00000162349.8 ENSMUST00000087256.12 ENSMUST00000160103.8 ENSMUST00000160870.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr17_+_38950335 | 0.55 |
ENSMUST00000174742.3
|
Esp31
|
exocrine gland secreted peptide 31 |
| chr7_+_55889488 | 0.53 |
ENSMUST00000032633.12
ENSMUST00000155533.2 ENSMUST00000156886.8 |
Oca2
|
oculocutaneous albinism II |
| chr11_-_77616103 | 0.50 |
ENSMUST00000078623.5
|
Cryba1
|
crystallin, beta A1 |
| chr13_+_120616163 | 0.50 |
ENSMUST00000179071.2
|
Gm20767
|
predicted gene, 20767 |
| chr6_-_42437951 | 0.49 |
ENSMUST00000090156.2
|
Olfr458
|
olfactory receptor 458 |
| chr17_+_38865083 | 0.49 |
ENSMUST00000173055.3
ENSMUST00000178654.2 |
Esp34
|
exocrine gland secreted peptide 34 |
| chrY_+_2599099 | 0.47 |
ENSMUST00000163651.2
|
H2al2c
|
H2A histone family member L2C |
| chr1_+_192984278 | 0.45 |
ENSMUST00000016315.16
|
Lamb3
|
laminin, beta 3 |
| chr13_+_120779103 | 0.45 |
ENSMUST00000178349.2
|
Tcstv3
|
2-cell-stage, variable group, member 3 |
| chr9_-_39694549 | 0.45 |
ENSMUST00000216458.3
|
Olfr968
|
olfactory receptor 968 |
| chr19_-_10502468 | 0.43 |
ENSMUST00000025570.8
ENSMUST00000236455.2 |
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr5_-_66776095 | 0.43 |
ENSMUST00000162366.8
ENSMUST00000162994.8 ENSMUST00000159512.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chrX_+_125947591 | 0.43 |
ENSMUST00000096332.2
|
Gm382
|
predicted gene 382 |
| chr13_+_23343482 | 0.41 |
ENSMUST00000226845.2
ENSMUST00000228666.2 ENSMUST00000227388.2 |
Vmn1r219
|
vomeronasal 1 receptor 219 |
| chr11_+_97206542 | 0.40 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr9_+_37714354 | 0.40 |
ENSMUST00000215287.2
|
Olfr876
|
olfactory receptor 876 |
| chr2_+_85838122 | 0.39 |
ENSMUST00000062166.2
|
Olfr1032
|
olfactory receptor 1032 |
| chr13_+_33508060 | 0.38 |
ENSMUST00000075515.7
|
Serpinb9f
|
serine (or cysteine) peptidase inhibitor, clade B, member 9f |
| chr5_-_26294289 | 0.35 |
ENSMUST00000094946.5
|
Gm10471
|
predicted gene 10471 |
| chr12_-_114579938 | 0.34 |
ENSMUST00000195469.6
ENSMUST00000109711.4 |
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
| chr2_-_30305401 | 0.33 |
ENSMUST00000142096.2
|
Crat
|
carnitine acetyltransferase |
| chr7_+_104963189 | 0.31 |
ENSMUST00000098153.3
|
Olfr689
|
olfactory receptor 689 |
| chr12_+_98886826 | 0.31 |
ENSMUST00000085109.10
ENSMUST00000079146.13 |
Ttc8
|
tetratricopeptide repeat domain 8 |
| chr14_+_51599213 | 0.29 |
ENSMUST00000162998.2
|
Gm7247
|
predicted gene 7247 |
| chr19_-_10502546 | 0.28 |
ENSMUST00000237827.2
|
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr19_-_12219023 | 0.28 |
ENSMUST00000087818.2
|
Olfr262
|
olfactory receptor 262 |
| chr8_-_70426910 | 0.28 |
ENSMUST00000116463.4
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr19_+_11863929 | 0.27 |
ENSMUST00000217281.2
|
Olfr1420
|
olfactory receptor 1420 |
| chr1_-_181841969 | 0.27 |
ENSMUST00000193074.6
|
Enah
|
ENAH actin regulator |
| chr7_+_104202091 | 0.26 |
ENSMUST00000098176.5
|
Olfr651
|
olfactory receptor 651 |
| chr5_+_91175323 | 0.25 |
ENSMUST00000202724.4
ENSMUST00000041516.9 |
Epgn
|
epithelial mitogen |
| chr3_+_101917455 | 0.24 |
ENSMUST00000066187.6
ENSMUST00000198675.2 |
Nhlh2
|
nescient helix loop helix 2 |
| chr10_-_78187545 | 0.24 |
ENSMUST00000105390.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr14_+_59716265 | 0.22 |
ENSMUST00000224893.2
|
Cab39l
|
calcium binding protein 39-like |
| chr11_-_78948432 | 0.20 |
ENSMUST00000129463.2
|
Ksr1
|
kinase suppressor of ras 1 |
| chr18_-_62729391 | 0.19 |
ENSMUST00000076194.6
|
Spink7
|
serine peptidase inhibitor, Kazal type 7 (putative) |
| chr2_-_88256367 | 0.19 |
ENSMUST00000102619.3
ENSMUST00000213190.2 |
Olfr1181
|
olfactory receptor 1181 |
| chr3_+_101917392 | 0.18 |
ENSMUST00000196324.2
|
Nhlh2
|
nescient helix loop helix 2 |
| chr10_-_78187524 | 0.18 |
ENSMUST00000166360.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr4_-_43840201 | 0.17 |
ENSMUST00000214281.2
|
Olfr157
|
olfactory receptor 157 |
| chr2_-_89030312 | 0.17 |
ENSMUST00000214709.2
ENSMUST00000215987.2 ENSMUST00000215562.2 ENSMUST00000220416.2 ENSMUST00000216445.2 ENSMUST00000217601.2 |
Olfr1226
|
olfactory receptor 1226 |
| chr10_+_112001581 | 0.16 |
ENSMUST00000170013.2
|
Caps2
|
calcyphosphine 2 |
| chr11_-_55042529 | 0.15 |
ENSMUST00000020502.9
ENSMUST00000069816.6 |
Slc36a3
|
solute carrier family 36 (proton/amino acid symporter), member 3 |
| chr1_+_24717722 | 0.10 |
ENSMUST00000186096.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr14_-_101846551 | 0.10 |
ENSMUST00000100340.4
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr19_-_11848669 | 0.05 |
ENSMUST00000087857.3
|
Olfr1419
|
olfactory receptor 1419 |
| chr8_+_71047010 | 0.04 |
ENSMUST00000211117.2
|
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr9_+_92339422 | 0.02 |
ENSMUST00000034941.9
|
Plscr4
|
phospholipid scramblase 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.4 | GO:0061206 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 2.2 | 6.7 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 1.8 | 5.5 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 1.4 | 4.3 | GO:0001762 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 1.2 | 3.7 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.1 | 9.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 1.1 | 4.4 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 1.1 | 4.3 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.9 | 3.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.9 | 2.7 | GO:0015898 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 0.8 | 3.2 | GO:0015744 | succinate transport(GO:0015744) |
| 0.8 | 9.0 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.7 | 4.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.7 | 2.0 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.7 | 4.7 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.6 | 3.0 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.5 | 3.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.5 | 1.6 | GO:2000724 | response to mineralocorticoid(GO:0051385) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.5 | 3.7 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.4 | 2.6 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.4 | 1.7 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.4 | 2.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.4 | 5.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.4 | 3.5 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 | 2.9 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.3 | 2.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.3 | 1.1 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.3 | 4.0 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.3 | 1.3 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.3 | 3.5 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 1.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 0.9 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 2.2 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.2 | 1.3 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 0.6 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.2 | 1.7 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 1.8 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.2 | 0.7 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 0.7 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 0.9 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 1.3 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.1 | 1.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.6 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.1 | 4.1 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.6 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.1 | 0.7 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.1 | 0.3 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 1.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 1.8 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.6 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 5.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 1.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 1.4 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 4.2 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 2.1 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.9 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.3 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 2.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 4.5 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.5 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 1.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 2.5 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.2 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.5 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.6 | 3.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.2 | 4.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 1.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 1.8 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 3.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 3.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 2.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 4.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 6.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 4.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 10.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 6.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 3.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 2.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 29.2 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.1 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 8.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0005369 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 1.2 | 4.7 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 1.2 | 3.5 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 1.1 | 3.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 1.0 | 5.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.9 | 2.7 | GO:0052598 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 0.8 | 5.5 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.7 | 5.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.7 | 4.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.6 | 4.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.6 | 1.8 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.5 | 1.6 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.4 | 2.6 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.4 | 9.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 2.0 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 0.7 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.3 | 3.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 6.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 3.7 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.3 | 4.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 4.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 9.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 3.0 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.2 | 3.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 2.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 2.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.7 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 3.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.0 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.1 | 1.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 3.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 1.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 9.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 2.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 2.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 9.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 8.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 4.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 2.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 4.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 9.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.3 | 4.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 4.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 9.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 2.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 3.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 4.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 4.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 3.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 5.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |