Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
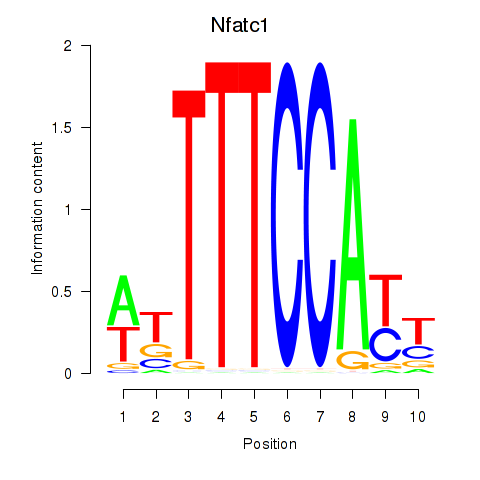
Results for Nfatc1
Z-value: 1.82
Transcription factors associated with Nfatc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc1
|
ENSMUSG00000033016.17 | Nfatc1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc1 | mm39_v1_chr18_-_80751327_80751395 | 0.74 | 1.7e-13 | Click! |
Activity profile of Nfatc1 motif
Sorted Z-values of Nfatc1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_88675700 | 11.62 |
ENSMUST00000087033.6
|
Jchain
|
immunoglobulin joining chain |
| chr2_+_35172392 | 11.56 |
ENSMUST00000028239.8
|
Gsn
|
gelsolin |
| chr17_-_48739874 | 10.92 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr3_-_106126794 | 10.38 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr15_-_41733099 | 10.25 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein |
| chr4_-_43523595 | 9.72 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr15_+_78810919 | 9.18 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr1_+_75336965 | 9.14 |
ENSMUST00000027409.10
|
Des
|
desmin |
| chr12_-_101784727 | 8.40 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chr15_-_83135920 | 8.10 |
ENSMUST00000229687.2
ENSMUST00000164614.3 ENSMUST00000049530.14 |
A4galt
|
alpha 1,4-galactosyltransferase |
| chr18_-_80756273 | 8.09 |
ENSMUST00000078049.12
ENSMUST00000236711.2 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr14_-_101846459 | 8.07 |
ENSMUST00000161991.8
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr8_+_46338498 | 8.02 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr3_-_151953894 | 7.88 |
ENSMUST00000196529.5
|
Nexn
|
nexilin |
| chr6_-_69792108 | 7.83 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr9_+_123902143 | 7.64 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr11_-_106641454 | 7.59 |
ENSMUST00000068021.9
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr8_+_46338557 | 7.46 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr14_+_79753055 | 7.17 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr1_-_66974492 | 7.11 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr1_-_66974694 | 7.08 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr13_-_41513215 | 7.08 |
ENSMUST00000224803.2
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9 |
| chr3_+_90520408 | 7.01 |
ENSMUST00000198128.2
|
S100a6
|
S100 calcium binding protein A6 (calcyclin) |
| chr10_+_115653152 | 7.01 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr19_-_34231600 | 6.97 |
ENSMUST00000238147.2
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr18_-_14105670 | 6.75 |
ENSMUST00000025288.9
|
Zfp521
|
zinc finger protein 521 |
| chr11_+_63023893 | 6.60 |
ENSMUST00000108700.2
|
Pmp22
|
peripheral myelin protein 22 |
| chr1_+_180158035 | 6.57 |
ENSMUST00000070181.7
|
Itpkb
|
inositol 1,4,5-trisphosphate 3-kinase B |
| chr11_+_63023395 | 6.48 |
ENSMUST00000108701.8
|
Pmp22
|
peripheral myelin protein 22 |
| chrX_+_72527208 | 6.44 |
ENSMUST00000033741.15
ENSMUST00000169489.2 |
Bgn
|
biglycan |
| chrX_+_158410229 | 6.39 |
ENSMUST00000112456.9
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr3_+_60436163 | 6.36 |
ENSMUST00000194201.6
ENSMUST00000193517.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr12_-_101785307 | 6.24 |
ENSMUST00000021603.9
|
Fbln5
|
fibulin 5 |
| chr3_-_151971391 | 6.13 |
ENSMUST00000199470.5
ENSMUST00000200589.5 |
Nexn
|
nexilin |
| chr6_+_38895902 | 6.08 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr7_-_126224848 | 6.06 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr3_+_90520176 | 6.01 |
ENSMUST00000001051.9
|
S100a6
|
S100 calcium binding protein A6 (calcyclin) |
| chr1_-_82746169 | 5.94 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr11_+_44508137 | 5.87 |
ENSMUST00000109268.2
ENSMUST00000101326.10 ENSMUST00000081265.12 |
Ebf1
|
early B cell factor 1 |
| chr10_+_33071964 | 5.79 |
ENSMUST00000219211.2
|
Trdn
|
triadin |
| chr4_-_43523745 | 5.76 |
ENSMUST00000150592.2
|
Tpm2
|
tropomyosin 2, beta |
| chr4_-_44710408 | 5.70 |
ENSMUST00000134968.9
ENSMUST00000173821.8 ENSMUST00000174319.8 ENSMUST00000173733.8 ENSMUST00000172866.8 ENSMUST00000165417.9 ENSMUST00000107825.9 ENSMUST00000102932.10 ENSMUST00000107827.9 ENSMUST00000107826.9 ENSMUST00000014174.14 |
Pax5
|
paired box 5 |
| chr11_-_75918551 | 5.66 |
ENSMUST00000021207.7
|
Rflnb
|
refilin B |
| chr6_+_41515152 | 5.66 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chr3_+_94284812 | 5.60 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr5_+_17779273 | 5.53 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr3_+_60436570 | 5.40 |
ENSMUST00000192607.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr3_+_94284739 | 5.34 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr12_-_114646685 | 5.34 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr2_-_73360069 | 5.30 |
ENSMUST00000094681.11
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chrX_+_106132055 | 5.28 |
ENSMUST00000150494.2
|
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr8_-_83128437 | 5.26 |
ENSMUST00000209573.3
|
Il15
|
interleukin 15 |
| chr1_-_134883645 | 5.26 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr10_-_27492827 | 5.23 |
ENSMUST00000092639.12
|
Lama2
|
laminin, alpha 2 |
| chr1_-_169938060 | 5.15 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr6_-_124865155 | 5.13 |
ENSMUST00000024044.7
|
Cd4
|
CD4 antigen |
| chrX_+_158410528 | 5.00 |
ENSMUST00000073094.10
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr15_-_66673425 | 4.98 |
ENSMUST00000168589.8
|
Sla
|
src-like adaptor |
| chrX_-_47543029 | 4.93 |
ENSMUST00000114958.8
|
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr1_+_107456731 | 4.91 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr6_+_41498716 | 4.84 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr4_-_129452148 | 4.83 |
ENSMUST00000167288.8
ENSMUST00000134336.3 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr4_-_129452180 | 4.81 |
ENSMUST00000067240.11
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr6_+_67586695 | 4.76 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr7_-_30520563 | 4.71 |
ENSMUST00000186339.7
ENSMUST00000163504.8 ENSMUST00000186059.2 |
Ffar2
|
free fatty acid receptor 2 |
| chr6_+_145879839 | 4.62 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr10_-_62343516 | 4.60 |
ENSMUST00000020271.13
|
Srgn
|
serglycin |
| chr9_+_123901979 | 4.51 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr11_+_95733489 | 4.51 |
ENSMUST00000100532.10
ENSMUST00000036088.11 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr11_-_83421333 | 4.49 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr7_+_100143250 | 4.47 |
ENSMUST00000153287.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr2_-_103133524 | 4.47 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor |
| chr2_-_103133503 | 4.46 |
ENSMUST00000111176.9
|
Ehf
|
ets homologous factor |
| chr3_-_145355725 | 4.44 |
ENSMUST00000029846.5
|
Ccn1
|
cellular communication network factor 1 |
| chr9_-_44437694 | 4.43 |
ENSMUST00000062215.8
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr12_-_114672701 | 4.42 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr11_-_100151847 | 4.34 |
ENSMUST00000080893.7
|
Krt17
|
keratin 17 |
| chr1_-_66984178 | 4.32 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr5_-_103777145 | 4.32 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr17_+_35643818 | 4.31 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr8_-_3928495 | 4.29 |
ENSMUST00000209176.2
ENSMUST00000011445.8 |
Cd209d
|
CD209d antigen |
| chr6_+_21986445 | 4.24 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr14_-_101846551 | 4.24 |
ENSMUST00000100340.4
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr13_+_83652352 | 4.24 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr17_+_35643853 | 4.19 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr8_+_107237483 | 4.18 |
ENSMUST00000080797.8
|
Cdh3
|
cadherin 3 |
| chr12_-_114710326 | 4.18 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr6_+_29060215 | 4.13 |
ENSMUST00000069789.12
|
Lep
|
leptin |
| chr16_+_45430743 | 4.12 |
ENSMUST00000161347.9
ENSMUST00000023339.5 |
Gcsam
|
germinal center associated, signaling and motility |
| chrX_-_9335525 | 4.09 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr4_-_141553306 | 4.09 |
ENSMUST00000102481.4
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr10_+_42736771 | 4.08 |
ENSMUST00000105494.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr4_-_84593226 | 4.00 |
ENSMUST00000175800.8
ENSMUST00000176418.8 ENSMUST00000175969.8 ENSMUST00000176370.2 ENSMUST00000176947.8 ENSMUST00000102820.9 ENSMUST00000107198.9 ENSMUST00000175756.8 ENSMUST00000176691.8 |
Bnc2
|
basonuclin 2 |
| chr6_+_70495224 | 3.89 |
ENSMUST00000103396.2
|
Igkv3-12
|
immunoglobulin kappa variable 3-12 |
| chr2_-_60793536 | 3.85 |
ENSMUST00000028347.13
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr7_+_100142977 | 3.84 |
ENSMUST00000129324.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr7_+_46496506 | 3.80 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr10_+_42736345 | 3.79 |
ENSMUST00000063063.14
|
Scml4
|
Scm polycomb group protein like 4 |
| chr1_-_95595280 | 3.78 |
ENSMUST00000043336.11
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr11_-_101785181 | 3.75 |
ENSMUST00000057054.8
|
Meox1
|
mesenchyme homeobox 1 |
| chr7_+_30252687 | 3.74 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr14_-_59578768 | 3.73 |
ENSMUST00000166121.4
|
Phf11b
|
PHD finger protein 11B |
| chr7_+_75292971 | 3.72 |
ENSMUST00000207998.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr4_-_87951565 | 3.71 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr15_+_61857390 | 3.71 |
ENSMUST00000159327.2
ENSMUST00000167731.8 |
Myc
|
myelocytomatosis oncogene |
| chr4_-_11386756 | 3.70 |
ENSMUST00000108313.8
ENSMUST00000108311.9 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr7_+_46496929 | 3.67 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr9_+_89081262 | 3.67 |
ENSMUST00000068569.5
|
Bcl2a1b
|
B cell leukemia/lymphoma 2 related protein A1b |
| chr17_+_87270504 | 3.66 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr19_-_57107330 | 3.65 |
ENSMUST00000111526.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr6_+_65502344 | 3.60 |
ENSMUST00000212375.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr1_+_133965228 | 3.59 |
ENSMUST00000162779.2
|
Fmod
|
fibromodulin |
| chr10_-_93146825 | 3.59 |
ENSMUST00000151153.2
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr3_+_95406567 | 3.58 |
ENSMUST00000015664.5
|
Ctsk
|
cathepsin K |
| chr11_+_44290884 | 3.57 |
ENSMUST00000170513.3
ENSMUST00000102796.10 |
Il12b
|
interleukin 12b |
| chr11_-_79421397 | 3.56 |
ENSMUST00000103236.4
ENSMUST00000170799.8 ENSMUST00000170422.4 |
Evi2a
Evi2
|
ecotropic viral integration site 2a ecotropic viral integration site 2 |
| chr13_-_100922910 | 3.55 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr13_-_117161921 | 3.54 |
ENSMUST00000223949.2
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr1_+_60948149 | 3.49 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr11_-_70545424 | 3.35 |
ENSMUST00000108549.2
|
Pfn1
|
profilin 1 |
| chr8_-_105350898 | 3.34 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr19_+_58658779 | 3.32 |
ENSMUST00000057270.9
|
Pnlip
|
pancreatic lipase |
| chr11_-_70545450 | 3.32 |
ENSMUST00000018437.3
|
Pfn1
|
profilin 1 |
| chrX_+_74425990 | 3.31 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr11_-_48836975 | 3.31 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr1_-_169938298 | 3.31 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr11_+_67090878 | 3.30 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr13_-_117162041 | 3.29 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr3_+_145827410 | 3.28 |
ENSMUST00000039450.5
|
Mcoln3
|
mucolipin 3 |
| chr2_+_18681812 | 3.27 |
ENSMUST00000028071.13
|
Bmi1
|
Bmi1 polycomb ring finger oncogene |
| chr1_-_170755136 | 3.27 |
ENSMUST00000046322.14
ENSMUST00000159171.2 |
Fcrla
|
Fc receptor-like A |
| chr9_-_123691077 | 3.26 |
ENSMUST00000182350.3
|
Xcr1
|
chemokine (C motif) receptor 1 |
| chr9_+_108183996 | 3.24 |
ENSMUST00000194701.6
ENSMUST00000193490.2 |
Rhoa
|
ras homolog family member A |
| chr14_+_22069709 | 3.24 |
ENSMUST00000075639.11
|
Lrmda
|
leucine rich melanocyte differentiation associated |
| chrX_+_163221035 | 3.20 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr7_-_104019046 | 3.17 |
ENSMUST00000106831.3
|
Trim30b
|
tripartite motif-containing 30B |
| chr4_-_11386679 | 3.14 |
ENSMUST00000043781.14
ENSMUST00000108310.8 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr15_-_56557920 | 3.09 |
ENSMUST00000050544.8
|
Has2
|
hyaluronan synthase 2 |
| chr6_+_65502292 | 3.07 |
ENSMUST00000212402.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr14_+_120513076 | 3.06 |
ENSMUST00000088419.13
|
Mbnl2
|
muscleblind like splicing factor 2 |
| chr15_+_6609322 | 3.06 |
ENSMUST00000090461.12
|
Fyb
|
FYN binding protein |
| chr10_-_18890281 | 3.05 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr11_-_78074377 | 3.00 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr3_+_87754057 | 3.00 |
ENSMUST00000107581.9
|
Sh2d2a
|
SH2 domain containing 2A |
| chr6_+_21985902 | 2.97 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr10_-_27492792 | 2.94 |
ENSMUST00000189575.2
|
Lama2
|
laminin, alpha 2 |
| chr11_-_100098333 | 2.93 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr6_+_149210941 | 2.93 |
ENSMUST00000190785.7
ENSMUST00000189932.7 ENSMUST00000100765.11 |
Resf1
|
retroelement silencing factor 1 |
| chr11_+_95733109 | 2.93 |
ENSMUST00000107714.9
ENSMUST00000107711.8 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr3_+_87754310 | 2.92 |
ENSMUST00000029709.7
|
Sh2d2a
|
SH2 domain containing 2A |
| chr6_+_97187650 | 2.92 |
ENSMUST00000044681.7
|
Arl6ip5
|
ADP-ribosylation factor-like 6 interacting protein 5 |
| chr17_+_37581103 | 2.92 |
ENSMUST00000038580.7
|
H2-M3
|
histocompatibility 2, M region locus 3 |
| chr11_+_81936531 | 2.91 |
ENSMUST00000021011.3
|
Ccl7
|
chemokine (C-C motif) ligand 7 |
| chr7_+_46496552 | 2.90 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr1_-_69726384 | 2.90 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr2_+_127750978 | 2.87 |
ENSMUST00000110344.2
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr7_-_104018989 | 2.86 |
ENSMUST00000164410.2
|
Trim30b
|
tripartite motif-containing 30B |
| chr2_-_181223787 | 2.86 |
ENSMUST00000057816.15
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chr1_-_134883577 | 2.86 |
ENSMUST00000168381.8
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr4_+_134591847 | 2.83 |
ENSMUST00000030627.8
|
Rhd
|
Rh blood group, D antigen |
| chr3_+_89979948 | 2.82 |
ENSMUST00000121503.8
ENSMUST00000119570.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr13_+_83652150 | 2.82 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr8_+_95161006 | 2.78 |
ENSMUST00000211816.2
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr9_+_69896748 | 2.76 |
ENSMUST00000034754.12
ENSMUST00000085393.13 ENSMUST00000117450.8 |
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr8_-_105350533 | 2.75 |
ENSMUST00000212662.2
|
Cdh16
|
cadherin 16 |
| chr3_-_135313982 | 2.74 |
ENSMUST00000132668.8
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr9_+_123635556 | 2.74 |
ENSMUST00000216072.2
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr9_+_123635529 | 2.73 |
ENSMUST00000049810.9
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chrX_-_12539778 | 2.70 |
ENSMUST00000060108.7
|
1810030O07Rik
|
RIKEN cDNA 1810030O07 gene |
| chr6_+_149210889 | 2.70 |
ENSMUST00000130664.8
ENSMUST00000046689.13 |
Resf1
|
retroelement silencing factor 1 |
| chr18_-_14105591 | 2.69 |
ENSMUST00000234025.2
|
Zfp521
|
zinc finger protein 521 |
| chr1_+_82210833 | 2.68 |
ENSMUST00000023262.6
|
Gm9747
|
predicted gene 9747 |
| chr5_+_67126304 | 2.67 |
ENSMUST00000132991.5
|
Limch1
|
LIM and calponin homology domains 1 |
| chr7_-_80475722 | 2.66 |
ENSMUST00000205304.2
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr8_-_105350881 | 2.65 |
ENSMUST00000211903.2
|
Cdh16
|
cadherin 16 |
| chr12_-_114878652 | 2.64 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr7_+_30122685 | 2.64 |
ENSMUST00000046177.9
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr11_+_96177449 | 2.62 |
ENSMUST00000049352.8
|
Hoxb7
|
homeobox B7 |
| chr16_-_21980200 | 2.60 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr19_-_47126719 | 2.59 |
ENSMUST00000140512.8
ENSMUST00000035822.2 |
Calhm2
|
calcium homeostasis modulator family member 2 |
| chr15_-_98505508 | 2.57 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr5_+_138228074 | 2.57 |
ENSMUST00000159798.8
ENSMUST00000159964.2 |
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr14_+_120513106 | 2.57 |
ENSMUST00000227012.2
ENSMUST00000167459.3 |
Mbnl2
|
muscleblind like splicing factor 2 |
| chr10_+_42736644 | 2.55 |
ENSMUST00000105495.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr3_-_101743539 | 2.52 |
ENSMUST00000061831.11
|
Mab21l3
|
mab-21-like 3 |
| chr7_+_27207226 | 2.52 |
ENSMUST00000125990.2
ENSMUST00000065487.7 |
Prx
|
periaxin |
| chr5_+_13449276 | 2.51 |
ENSMUST00000030714.9
ENSMUST00000141968.2 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr10_+_67021509 | 2.49 |
ENSMUST00000173689.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chr1_-_131161312 | 2.45 |
ENSMUST00000212202.2
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr10_-_93146937 | 2.44 |
ENSMUST00000008542.12
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr9_-_58065800 | 2.43 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr19_+_58658838 | 2.42 |
ENSMUST00000238108.2
|
Pnlip
|
pancreatic lipase |
| chr6_+_70584375 | 2.42 |
ENSMUST00000197560.2
|
Igkv3-7
|
immunoglobulin kappa variable 3-7 |
| chr3_-_57483175 | 2.38 |
ENSMUST00000029380.14
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr12_+_75355082 | 2.38 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr5_+_3393893 | 2.37 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr7_+_88079465 | 2.36 |
ENSMUST00000107256.4
|
Rab38
|
RAB38, member RAS oncogene family |
| chr7_+_27770655 | 2.29 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr15_+_101152078 | 2.29 |
ENSMUST00000228985.2
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_-_170755109 | 2.29 |
ENSMUST00000162136.2
ENSMUST00000162887.2 |
Fcrla
|
Fc receptor-like A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 3.1 | 9.2 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 2.9 | 11.6 | GO:1903921 | renal protein absorption(GO:0097017) protein processing in phagocytic vesicle(GO:1900756) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 2.7 | 8.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 2.5 | 7.6 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 2.2 | 4.5 | GO:0035697 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) |
| 2.1 | 8.5 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.7 | 10.4 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 1.7 | 6.9 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 1.4 | 8.5 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 1.4 | 4.2 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 1.4 | 4.1 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 1.4 | 8.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 1.3 | 15.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.3 | 2.6 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 1.3 | 1.3 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 1.2 | 11.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.2 | 12.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 1.2 | 7.0 | GO:0035120 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.2 | 4.6 | GO:0033380 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 1.1 | 3.4 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 1.1 | 6.6 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 1.1 | 3.3 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 1.0 | 10.4 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.0 | 3.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 1.0 | 12.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 1.0 | 3.1 | GO:0002631 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 1.0 | 3.0 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 1.0 | 6.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 1.0 | 10.9 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 1.0 | 2.9 | GO:0002481 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.9 | 4.7 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.9 | 3.7 | GO:0090095 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.9 | 11.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.9 | 2.7 | GO:1904629 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.9 | 5.5 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.9 | 8.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.9 | 3.6 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.9 | 7.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.8 | 10.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.8 | 3.2 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.8 | 2.4 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.7 | 8.2 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.7 | 9.6 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.7 | 2.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.7 | 2.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.7 | 2.0 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.7 | 2.0 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.7 | 2.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.6 | 14.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.6 | 3.8 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.6 | 3.7 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.6 | 4.3 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.6 | 4.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.6 | 2.9 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.6 | 2.3 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.5 | 8.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.5 | 6.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.5 | 4.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.5 | 1.5 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.5 | 16.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.5 | 1.9 | GO:0042335 | cuticle development(GO:0042335) |
| 0.5 | 1.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.5 | 2.4 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.5 | 1.9 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.5 | 1.9 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.4 | 2.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.4 | 2.6 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.4 | 8.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.4 | 13.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 2.5 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.4 | 1.7 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.4 | 2.9 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.4 | 4.1 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.4 | 1.6 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.4 | 2.8 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.4 | 3.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.4 | 6.1 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.4 | 15.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.4 | 6.7 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.3 | 4.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 2.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.3 | 6.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.3 | 5.7 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.3 | 2.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.3 | 1.2 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.3 | 2.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.3 | 3.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 1.7 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.3 | 2.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 1.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 0.8 | GO:0097184 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 0.3 | 0.8 | GO:0048880 | medulla oblongata development(GO:0021550) hindbrain tangential cell migration(GO:0021934) sensory system development(GO:0048880) |
| 0.3 | 5.7 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.3 | 1.0 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.3 | 13.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.3 | 4.8 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.2 | 1.7 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 0.5 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 1.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 2.7 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.2 | 2.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 1.3 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 | 1.8 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.2 | 8.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.2 | 2.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 6.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.2 | 0.6 | GO:1904735 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.2 | 7.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 1.2 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 7.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 0.6 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.2 | 1.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.2 | 4.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 1.3 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.2 | 1.3 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.2 | 1.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.2 | 6.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.2 | 23.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 0.6 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 7.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 | 1.5 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 1.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 0.6 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 5.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.7 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 2.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 3.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 1.5 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 1.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 3.6 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 1.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 2.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 3.7 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 1.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 1.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 2.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.9 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 3.7 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.1 | 1.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 1.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 2.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 5.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.8 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 1.0 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) positive regulation of mononuclear cell migration(GO:0071677) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 3.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.5 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 4.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 3.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.5 | GO:0044857 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 1.7 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 1.4 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.9 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.1 | 1.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 1.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 5.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 13.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 1.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 3.3 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 1.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 1.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 3.7 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 1.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 12.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.8 | GO:0043276 | anoikis(GO:0043276) |
| 0.1 | 1.1 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.6 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 3.5 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 1.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 3.1 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 3.7 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 1.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.9 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 1.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 1.5 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 2.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.1 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 6.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 2.9 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 1.8 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.5 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.7 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 3.1 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 1.5 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 2.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.4 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.5 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 2.2 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0070895 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.6 | 14.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.4 | 5.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.2 | 18.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.2 | 7.0 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 1.2 | 11.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.9 | 3.6 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.8 | 9.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.8 | 2.4 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.7 | 2.8 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.6 | 8.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.5 | 4.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 4.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 13.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.4 | 10.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.4 | 10.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 3.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.6 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.3 | 1.6 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.3 | 4.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 1.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 0.9 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.3 | 35.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 7.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 8.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 4.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 3.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 0.7 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 5.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 2.9 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.2 | 1.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 2.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 2.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 1.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 21.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 3.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 2.9 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 0.5 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 12.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 37.2 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 15.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 10.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 2.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 24.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 5.5 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 3.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.8 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 5.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 5.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 6.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.3 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 3.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 9.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 7.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 6.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 23.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 34.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 16.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 7.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 4.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 6.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 31.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 3.9 | 15.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 2.9 | 11.6 | GO:0019862 | IgA binding(GO:0019862) |
| 2.3 | 9.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.9 | 9.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.8 | 10.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.6 | 8.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 1.6 | 9.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.6 | 4.7 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.5 | 6.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.5 | 4.5 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 1.4 | 4.2 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 1.4 | 8.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.2 | 11.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.2 | 10.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 1.0 | 3.0 | GO:0005368 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 1.0 | 7.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.0 | 11.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.9 | 10.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.9 | 11.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.8 | 18.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.8 | 3.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.8 | 8.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.7 | 2.9 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.7 | 6.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.7 | 2.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.7 | 47.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.7 | 2.0 | GO:0016872 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 0.6 | 6.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.6 | 3.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.6 | 2.4 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.6 | 1.7 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.5 | 8.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.5 | 1.9 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.4 | 3.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 8.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 2.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 11.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 2.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.4 | 8.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 1.2 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.4 | 1.6 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.4 | 3.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 6.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 1.1 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.4 | 2.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.4 | 3.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 1.4 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.3 | 2.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 4.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 0.9 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 2.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 5.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.3 | 0.8 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.3 | 1.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.3 | 0.8 | GO:0052600 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 0.3 | 5.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 3.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.3 | 1.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.3 | 5.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 0.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 0.7 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 2.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 6.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 1.3 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 3.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 5.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 23.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 5.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 10.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 4.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 3.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 2.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.2 | 1.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 1.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 3.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 2.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 5.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.4 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.1 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 10.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.6 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 3.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 2.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 18.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 1.0 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 6.1 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.1 | 3.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 4.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 3.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.9 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 3.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 6.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.5 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 2.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 3.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.5 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 2.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 2.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 6.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.0 | 1.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 31.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 7.6 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 1.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 14.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 5.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.9 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 3.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 11.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 4.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.2 | GO:0001848 | complement binding(GO:0001848) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.5 | 8.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 16.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.4 | 14.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.3 | 11.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 15.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 12.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 14.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 8.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 7.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 5.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 7.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 11.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 9.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 11.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 7.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 16.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 2.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 7.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 7.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 8.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 5.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 2.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 5.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 4.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 5.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 15.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 17.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 0.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 2.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 12.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 5.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 5.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 5.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 4.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 9.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 22.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 1.0 | 9.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 46.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 23.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.5 | 6.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 5.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 10.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.4 | 11.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 4.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 7.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.3 | 13.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.3 | 5.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 5.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 17.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 3.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 7.0 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 4.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 7.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.2 | 5.5 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.2 | 7.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 3.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 2.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 6.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 2.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 2.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 3.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 6.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 6.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 5.7 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 2.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 8.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 3.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 8.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 1.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 7.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 5.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 3.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 2.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 7.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 7.5 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 3.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 3.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.2 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |