Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
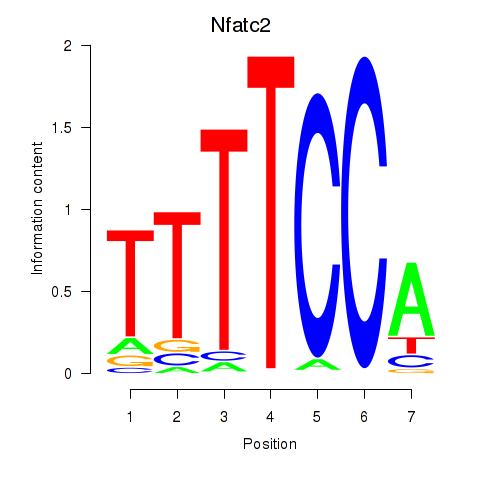
Results for Nfatc2
Z-value: 1.52
Transcription factors associated with Nfatc2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc2
|
ENSMUSG00000027544.17 | Nfatc2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc2 | mm39_v1_chr2_-_168443540_168443577 | 0.31 | 8.2e-03 | Click! |
Activity profile of Nfatc2 motif
Sorted Z-values of Nfatc2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_60229164 | 20.76 |
ENSMUST00000166381.3
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr2_-_52448552 | 7.45 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr6_-_36787096 | 7.06 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr16_+_41353360 | 6.78 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr17_-_91396154 | 6.56 |
ENSMUST00000161402.10
ENSMUST00000054059.15 ENSMUST00000072671.14 ENSMUST00000174331.8 |
Nrxn1
|
neurexin I |
| chrX_+_165127688 | 6.32 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr6_-_124746510 | 6.24 |
ENSMUST00000149652.2
ENSMUST00000112476.8 ENSMUST00000004378.15 |
Eno2
|
enolase 2, gamma neuronal |
| chr5_+_31011140 | 6.03 |
ENSMUST00000202501.2
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr2_+_3115250 | 5.97 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr10_+_76089674 | 5.44 |
ENSMUST00000036387.8
|
S100b
|
S100 protein, beta polypeptide, neural |
| chr2_+_121188195 | 5.41 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr17_+_24026892 | 5.34 |
ENSMUST00000191385.3
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr2_+_121697398 | 5.06 |
ENSMUST00000110586.10
ENSMUST00000078752.10 |
Golm2
|
golgi membrane protein 2 |
| chr12_-_72283465 | 4.99 |
ENSMUST00000021497.16
ENSMUST00000137990.2 |
Rtn1
|
reticulon 1 |
| chr4_+_102617495 | 4.74 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr18_+_37818263 | 4.66 |
ENSMUST00000194418.2
|
Pcdhga4
|
protocadherin gamma subfamily A, 4 |
| chr2_-_79959178 | 4.60 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr9_-_75504926 | 4.60 |
ENSMUST00000164100.2
|
Tmod2
|
tropomodulin 2 |
| chr19_-_11848669 | 4.58 |
ENSMUST00000087857.3
|
Olfr1419
|
olfactory receptor 1419 |
| chr7_+_130179063 | 4.58 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr5_+_16139760 | 4.53 |
ENSMUST00000101581.10
ENSMUST00000039370.14 ENSMUST00000199704.5 ENSMUST00000180204.8 ENSMUST00000078272.13 ENSMUST00000115281.7 |
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr12_+_76451177 | 4.52 |
ENSMUST00000219555.2
|
Hspa2
|
heat shock protein 2 |
| chrX_-_94209913 | 4.52 |
ENSMUST00000113873.9
ENSMUST00000113876.9 ENSMUST00000199920.5 ENSMUST00000113885.8 ENSMUST00000113883.8 ENSMUST00000196012.2 ENSMUST00000182001.8 ENSMUST00000113878.8 ENSMUST00000113882.8 ENSMUST00000182562.2 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr8_-_65146079 | 4.51 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr14_+_76192449 | 4.51 |
ENSMUST00000050120.4
|
Kctd4
|
potassium channel tetramerisation domain containing 4 |
| chr12_-_11486544 | 4.46 |
ENSMUST00000072299.7
|
Vsnl1
|
visinin-like 1 |
| chr10_-_12689345 | 4.42 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr10_-_127098932 | 4.31 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr7_-_126399208 | 4.24 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr18_+_37875135 | 4.24 |
ENSMUST00000003599.9
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr18_+_37813286 | 4.23 |
ENSMUST00000192931.2
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1 |
| chr16_+_13804461 | 4.21 |
ENSMUST00000056521.12
ENSMUST00000118412.8 ENSMUST00000131608.2 |
Bmerb1
|
bMERB domain containing 1 |
| chr3_-_33137165 | 4.21 |
ENSMUST00000078226.10
ENSMUST00000108224.8 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr16_+_41353212 | 4.21 |
ENSMUST00000078873.11
|
Lsamp
|
limbic system-associated membrane protein |
| chr6_+_120750510 | 4.17 |
ENSMUST00000112682.4
|
Slc25a18
|
solute carrier family 25 (mitochondrial carrier), member 18 |
| chr3_-_116762617 | 4.11 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr1_-_65162267 | 4.08 |
ENSMUST00000050047.4
ENSMUST00000148020.8 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr18_-_15284476 | 4.06 |
ENSMUST00000025992.7
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr5_+_16139683 | 4.04 |
ENSMUST00000167946.9
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr6_-_18514801 | 3.96 |
ENSMUST00000090601.12
|
Cttnbp2
|
cortactin binding protein 2 |
| chr11_+_102727122 | 3.93 |
ENSMUST00000021302.15
ENSMUST00000107072.2 |
Higd1b
|
HIG1 domain family, member 1B |
| chr12_-_90705212 | 3.87 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chrX_-_16777913 | 3.85 |
ENSMUST00000040134.8
|
Ndp
|
Norrie disease (pseudoglioma) (human) |
| chr6_+_90443293 | 3.83 |
ENSMUST00000203607.2
|
Klf15
|
Kruppel-like factor 15 |
| chr5_+_17779273 | 3.81 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr19_-_8816530 | 3.81 |
ENSMUST00000096259.6
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr15_+_78314251 | 3.79 |
ENSMUST00000229622.2
ENSMUST00000162808.2 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr12_+_103564479 | 3.79 |
ENSMUST00000190151.2
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr2_+_157401998 | 3.76 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr11_-_99313078 | 3.73 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr11_+_96177449 | 3.68 |
ENSMUST00000049352.8
|
Hoxb7
|
homeobox B7 |
| chr11_-_35871300 | 3.63 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr13_-_113182891 | 3.61 |
ENSMUST00000231962.2
ENSMUST00000022282.6 |
Gpx8
|
glutathione peroxidase 8 (putative) |
| chr18_+_37143758 | 3.60 |
ENSMUST00000115657.10
ENSMUST00000192447.6 |
Pcdha11
|
protocadherin alpha 11 |
| chr9_-_95632387 | 3.57 |
ENSMUST00000189137.7
ENSMUST00000053785.10 |
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr15_-_57128522 | 3.55 |
ENSMUST00000137764.2
ENSMUST00000022995.13 |
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr18_+_37072232 | 3.55 |
ENSMUST00000115662.9
ENSMUST00000195590.2 |
Pcdha2
|
protocadherin alpha 2 |
| chr18_-_60781365 | 3.54 |
ENSMUST00000143275.3
|
Synpo
|
synaptopodin |
| chrX_+_152506577 | 3.52 |
ENSMUST00000140575.8
ENSMUST00000208373.2 ENSMUST00000185492.7 ENSMUST00000149514.8 |
Nbdy
|
negative regulator of P-body association |
| chr3_+_159545309 | 3.51 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr2_+_109522781 | 3.50 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr15_-_77037972 | 3.46 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr7_-_101570382 | 3.45 |
ENSMUST00000098236.9
|
Lrrc51
|
leucine rich repeat containing 51 |
| chr1_-_172125555 | 3.45 |
ENSMUST00000085913.11
ENSMUST00000097464.4 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr15_+_91557378 | 3.43 |
ENSMUST00000060642.7
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr17_+_12803019 | 3.42 |
ENSMUST00000046959.9
ENSMUST00000233066.2 |
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr11_-_98666159 | 3.42 |
ENSMUST00000064941.7
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr5_+_67126304 | 3.41 |
ENSMUST00000132991.5
|
Limch1
|
LIM and calponin homology domains 1 |
| chr3_-_33137209 | 3.40 |
ENSMUST00000194016.6
ENSMUST00000193681.6 ENSMUST00000192093.6 ENSMUST00000193289.6 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr9_+_113641615 | 3.38 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr5_+_117501557 | 3.36 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr1_-_75254989 | 3.35 |
ENSMUST00000039534.11
|
Resp18
|
regulated endocrine-specific protein 18 |
| chr8_-_105350898 | 3.35 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr18_-_31450095 | 3.35 |
ENSMUST00000139924.2
ENSMUST00000153060.8 |
Rit2
|
Ras-like without CAAX 2 |
| chr18_+_37085673 | 3.34 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr5_-_35259847 | 3.30 |
ENSMUST00000153664.2
|
Lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr9_+_37278647 | 3.30 |
ENSMUST00000051839.9
|
Hepacam
|
hepatocyte cell adhesion molecule |
| chr5_-_70999547 | 3.28 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr6_-_21851827 | 3.27 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr14_+_115329676 | 3.24 |
ENSMUST00000176912.8
ENSMUST00000175665.8 |
Gpc5
|
glypican 5 |
| chr1_-_91386976 | 3.24 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2 |
| chr6_-_118432436 | 3.23 |
ENSMUST00000161519.8
ENSMUST00000069292.14 |
Zfp248
|
zinc finger protein 248 |
| chr7_+_107166653 | 3.23 |
ENSMUST00000120990.2
|
Olfml1
|
olfactomedin-like 1 |
| chr14_-_47059546 | 3.22 |
ENSMUST00000226937.2
|
Gmfb
|
glia maturation factor, beta |
| chr13_-_19803928 | 3.19 |
ENSMUST00000221014.2
ENSMUST00000002885.8 ENSMUST00000220944.2 |
Epdr1
|
ependymin related protein 1 (zebrafish) |
| chr16_+_94225942 | 3.19 |
ENSMUST00000141176.2
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr15_-_79048674 | 3.18 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chr1_-_55265925 | 3.16 |
ENSMUST00000027121.15
ENSMUST00000114428.3 |
Rftn2
|
raftlin family member 2 |
| chrX_+_10351360 | 3.16 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr12_+_55883101 | 3.15 |
ENSMUST00000059250.8
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr11_-_79394904 | 3.11 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr5_-_131645437 | 3.11 |
ENSMUST00000161804.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr10_-_29411857 | 3.09 |
ENSMUST00000092623.5
|
Rspo3
|
R-spondin 3 |
| chr11_-_42070517 | 3.07 |
ENSMUST00000206105.2
ENSMUST00000153147.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr9_-_54554483 | 3.07 |
ENSMUST00000128163.8
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr3_+_8574420 | 3.05 |
ENSMUST00000029002.9
|
Stmn2
|
stathmin-like 2 |
| chr7_-_101570393 | 3.02 |
ENSMUST00000106965.8
ENSMUST00000106968.8 ENSMUST00000106967.8 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr12_+_75355082 | 3.01 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr10_-_20600442 | 3.00 |
ENSMUST00000170265.8
|
Pde7b
|
phosphodiesterase 7B |
| chr3_-_57202546 | 2.97 |
ENSMUST00000196506.2
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr14_-_20027279 | 2.97 |
ENSMUST00000160013.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr7_-_89176294 | 2.91 |
ENSMUST00000207932.2
|
Prss23
|
protease, serine 23 |
| chr5_-_5315968 | 2.90 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr3_-_95789505 | 2.90 |
ENSMUST00000159863.2
ENSMUST00000159739.8 ENSMUST00000036418.10 ENSMUST00000161866.8 |
Ciart
|
circadian associated repressor of transcription |
| chr5_-_71815318 | 2.88 |
ENSMUST00000199357.2
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr2_+_61634797 | 2.85 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
| chr3_+_7568481 | 2.82 |
ENSMUST00000051064.9
ENSMUST00000193010.2 |
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr13_+_93441447 | 2.79 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr19_+_4761181 | 2.79 |
ENSMUST00000008991.8
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr1_-_131995661 | 2.78 |
ENSMUST00000046658.10
|
Mfsd4a
|
major facilitator superfamily domain containing 4A |
| chr1_+_179928709 | 2.78 |
ENSMUST00000133890.8
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr17_-_49871291 | 2.78 |
ENSMUST00000224595.2
ENSMUST00000057610.8 |
Daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr11_+_60956624 | 2.77 |
ENSMUST00000041944.9
ENSMUST00000108717.3 |
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr10_-_108846816 | 2.76 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr3_-_84167119 | 2.75 |
ENSMUST00000107691.8
|
Trim2
|
tripartite motif-containing 2 |
| chr8_-_105350881 | 2.75 |
ENSMUST00000211903.2
|
Cdh16
|
cadherin 16 |
| chr8_-_37200051 | 2.73 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chrX_-_23151771 | 2.73 |
ENSMUST00000115319.9
|
Klhl13
|
kelch-like 13 |
| chr5_+_67473069 | 2.71 |
ENSMUST00000202770.2
|
Slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr12_+_84161095 | 2.71 |
ENSMUST00000123491.8
ENSMUST00000046340.9 ENSMUST00000136159.2 |
Dnal1
|
dynein, axonemal, light chain 1 |
| chr10_+_38841511 | 2.71 |
ENSMUST00000019992.6
|
Lama4
|
laminin, alpha 4 |
| chr18_+_43340616 | 2.68 |
ENSMUST00000045477.6
|
Stk32a
|
serine/threonine kinase 32A |
| chr5_-_121148143 | 2.68 |
ENSMUST00000202406.4
ENSMUST00000200792.2 |
Rph3a
|
rabphilin 3A |
| chr4_-_122854966 | 2.68 |
ENSMUST00000030408.12
ENSMUST00000127047.2 |
Mfsd2a
|
major facilitator superfamily domain containing 2A |
| chr13_+_49761506 | 2.68 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr4_+_19818718 | 2.67 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr3_+_64884839 | 2.67 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr18_+_37840092 | 2.65 |
ENSMUST00000195823.2
|
Pcdhga6
|
protocadherin gamma subfamily A, 6 |
| chr9_-_40366966 | 2.63 |
ENSMUST00000165104.8
ENSMUST00000045682.7 |
Gramd1b
|
GRAM domain containing 1B |
| chr1_-_156032568 | 2.63 |
ENSMUST00000015628.4
|
Fam163a
|
family with sequence similarity 163, member A |
| chr2_-_115896279 | 2.62 |
ENSMUST00000110907.8
ENSMUST00000110908.9 |
Meis2
|
Meis homeobox 2 |
| chr3_-_88679881 | 2.62 |
ENSMUST00000090945.5
|
Syt11
|
synaptotagmin XI |
| chr19_+_6547790 | 2.61 |
ENSMUST00000113458.8
ENSMUST00000113459.2 |
Nrxn2
|
neurexin II |
| chr16_+_7011580 | 2.60 |
ENSMUST00000231194.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr3_+_129974531 | 2.59 |
ENSMUST00000080335.11
ENSMUST00000106353.2 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr14_+_69266566 | 2.59 |
ENSMUST00000014957.10
|
Stc1
|
stanniocalcin 1 |
| chr14_-_20027219 | 2.58 |
ENSMUST00000055100.14
ENSMUST00000162425.8 |
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr6_-_127746390 | 2.58 |
ENSMUST00000032500.9
|
Prmt8
|
protein arginine N-methyltransferase 8 |
| chr5_-_44956932 | 2.57 |
ENSMUST00000199261.2
ENSMUST00000199534.5 |
Ldb2
|
LIM domain binding 2 |
| chr13_+_42834039 | 2.56 |
ENSMUST00000128646.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr14_+_52222283 | 2.56 |
ENSMUST00000093813.12
ENSMUST00000100639.11 ENSMUST00000182909.8 ENSMUST00000182760.8 ENSMUST00000182061.8 ENSMUST00000182193.2 |
Arhgef40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr1_+_171077984 | 2.55 |
ENSMUST00000111315.9
ENSMUST00000219033.2 |
Adamts4
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 4 |
| chr6_+_129510145 | 2.54 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr5_+_53966956 | 2.53 |
ENSMUST00000037337.10
|
Tbc1d19
|
TBC1 domain family, member 19 |
| chr16_-_56706494 | 2.52 |
ENSMUST00000023435.6
|
Tmem45a
|
transmembrane protein 45a |
| chr2_-_115895202 | 2.52 |
ENSMUST00000110906.9
|
Meis2
|
Meis homeobox 2 |
| chr9_+_77848556 | 2.52 |
ENSMUST00000134072.2
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr3_-_33898405 | 2.51 |
ENSMUST00000029222.8
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr5_-_112542671 | 2.51 |
ENSMUST00000196256.2
|
Asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr12_-_112477536 | 2.51 |
ENSMUST00000066791.7
|
Tmem179
|
transmembrane protein 179 |
| chr19_+_6434416 | 2.50 |
ENSMUST00000035269.15
ENSMUST00000113483.2 |
Pygm
|
muscle glycogen phosphorylase |
| chr5_-_118382926 | 2.50 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr15_+_10952418 | 2.50 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr3_+_113824181 | 2.50 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr10_+_3316505 | 2.48 |
ENSMUST00000217573.2
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr6_-_121450547 | 2.47 |
ENSMUST00000046373.8
ENSMUST00000151397.3 |
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr2_-_115895528 | 2.47 |
ENSMUST00000028639.13
ENSMUST00000102538.11 |
Meis2
|
Meis homeobox 2 |
| chr1_-_130656985 | 2.47 |
ENSMUST00000189534.7
|
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chrX_-_74621828 | 2.46 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr14_-_70864448 | 2.46 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chrX_-_142176889 | 2.45 |
ENSMUST00000166406.3
ENSMUST00000207415.2 |
Chrdl1
|
chordin-like 1 |
| chr1_+_179938904 | 2.45 |
ENSMUST00000145181.2
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr7_+_83234118 | 2.44 |
ENSMUST00000039317.14
ENSMUST00000164944.2 |
Tmc3
|
transmembrane channel-like gene family 3 |
| chr12_+_76450941 | 2.44 |
ENSMUST00000080449.7
|
Hspa2
|
heat shock protein 2 |
| chr8_+_63404228 | 2.44 |
ENSMUST00000118003.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr4_+_129941633 | 2.43 |
ENSMUST00000044565.15
ENSMUST00000132251.2 |
Col16a1
|
collagen, type XVI, alpha 1 |
| chr9_-_58065800 | 2.42 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr14_-_20027112 | 2.42 |
ENSMUST00000159028.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr16_-_34083549 | 2.41 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr15_-_44651411 | 2.41 |
ENSMUST00000090057.6
ENSMUST00000110269.8 ENSMUST00000228639.2 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chrX_+_73352694 | 2.39 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr5_-_24806960 | 2.38 |
ENSMUST00000030791.12
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr11_-_3864664 | 2.38 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr4_+_102617332 | 2.38 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr7_-_100306160 | 2.37 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr18_+_37898633 | 2.36 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr7_+_91321500 | 2.34 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr2_-_115894993 | 2.32 |
ENSMUST00000074285.8
|
Meis2
|
Meis homeobox 2 |
| chr14_+_59716265 | 2.32 |
ENSMUST00000224893.2
|
Cab39l
|
calcium binding protein 39-like |
| chr19_-_6593049 | 2.31 |
ENSMUST00000113451.9
|
Slc22a12
|
solute carrier family 22 (organic anion/cation transporter), member 12 |
| chr2_+_85606930 | 2.31 |
ENSMUST00000076250.2
|
Olfr1014
|
olfactory receptor 1014 |
| chr10_-_8632519 | 2.31 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr10_+_116111441 | 2.31 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr11_-_97591150 | 2.30 |
ENSMUST00000018681.14
|
Pcgf2
|
polycomb group ring finger 2 |
| chr16_-_20440005 | 2.30 |
ENSMUST00000052939.4
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr7_+_126396779 | 2.30 |
ENSMUST00000205324.2
|
Tlcd3b
|
TLC domain containing 3B |
| chr1_-_186438177 | 2.30 |
ENSMUST00000045288.14
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr7_+_134272395 | 2.29 |
ENSMUST00000211593.2
ENSMUST00000084488.5 |
Dock1
|
dedicator of cytokinesis 1 |
| chr19_+_25384024 | 2.29 |
ENSMUST00000146647.3
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr13_+_93441307 | 2.28 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr9_-_29874401 | 2.28 |
ENSMUST00000075069.11
|
Ntm
|
neurotrimin |
| chr7_-_65177444 | 2.28 |
ENSMUST00000206228.2
|
Tjp1
|
tight junction protein 1 |
| chr15_-_98851566 | 2.28 |
ENSMUST00000097014.7
|
Tuba1a
|
tubulin, alpha 1A |
| chr2_+_21372338 | 2.26 |
ENSMUST00000055946.8
|
Gpr158
|
G protein-coupled receptor 158 |
| chr1_+_159351337 | 2.26 |
ENSMUST00000192069.6
|
Tnr
|
tenascin R |
| chrX_+_93679671 | 2.26 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr2_+_61876806 | 2.25 |
ENSMUST00000102735.10
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr15_-_98193995 | 2.25 |
ENSMUST00000023722.12
ENSMUST00000169721.3 |
Zfp641
|
zinc finger protein 641 |
| chrX_-_74918122 | 2.25 |
ENSMUST00000033547.14
|
Pls3
|
plastin 3 (T-isoform) |
| chr5_+_89175815 | 2.24 |
ENSMUST00000130041.8
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 1.9 | 9.6 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 1.8 | 7.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.5 | 6.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 1.5 | 4.5 | GO:0030070 | insulin processing(GO:0030070) |
| 1.5 | 2.9 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 1.4 | 1.4 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 1.2 | 3.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.2 | 3.5 | GO:0061193 | taste bud development(GO:0061193) |
| 1.1 | 5.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 1.1 | 8.8 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 1.1 | 4.4 | GO:0061235 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 1.1 | 6.6 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 1.1 | 3.2 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 1.1 | 1.1 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 1.0 | 3.9 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 1.0 | 3.9 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 1.0 | 3.8 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.9 | 3.8 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.9 | 3.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.9 | 2.7 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 0.9 | 4.4 | GO:1905161 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.9 | 4.3 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.8 | 4.9 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.8 | 6.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.8 | 3.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.8 | 2.3 | GO:1905006 | positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.8 | 2.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.8 | 2.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.7 | 3.7 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.7 | 2.2 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.7 | 5.8 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.7 | 2.9 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.7 | 3.5 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.7 | 1.4 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.7 | 3.5 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.7 | 4.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.7 | 2.1 | GO:1904268 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.7 | 2.1 | GO:2000184 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.7 | 2.1 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.7 | 3.4 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.7 | 3.3 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.7 | 19.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.6 | 4.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.6 | 8.7 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.6 | 1.8 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.6 | 4.2 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.6 | 7.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.6 | 17.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.6 | 8.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.6 | 1.7 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.6 | 7.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.6 | 2.9 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.6 | 1.7 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.6 | 3.3 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.5 | 4.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.5 | 3.8 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.5 | 2.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.5 | 3.8 | GO:0035989 | tendon development(GO:0035989) |
| 0.5 | 3.2 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.5 | 1.6 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.5 | 2.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.5 | 1.6 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.5 | 4.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.5 | 1.5 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.5 | 6.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.5 | 2.9 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.5 | 3.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.5 | 1.4 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.5 | 12.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.5 | 1.8 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.5 | 0.9 | GO:0072240 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.5 | 3.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 5.8 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.4 | 1.3 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.4 | 2.7 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.4 | 2.6 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.4 | 1.3 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.4 | 0.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.4 | 3.7 | GO:0046959 | habituation(GO:0046959) |
| 0.4 | 1.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.4 | 3.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 3.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.4 | 6.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 1.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.4 | 4.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 3.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.4 | 2.6 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.4 | 1.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.4 | 1.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.4 | 2.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.4 | 1.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.4 | 2.5 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.4 | 7.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 1.7 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 1.4 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.3 | 2.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 1.4 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.3 | 1.7 | GO:0042631 | glycerol transport(GO:0015793) cellular response to water deprivation(GO:0042631) |
| 0.3 | 2.4 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 3.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 0.7 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.3 | 1.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.3 | 4.0 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 1.3 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.3 | 1.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.3 | 3.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.3 | 2.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.3 | 5.1 | GO:1903392 | negative regulation of adherens junction organization(GO:1903392) |
| 0.3 | 2.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 1.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.3 | 7.5 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.3 | 1.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 0.6 | GO:0071317 | cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.3 | 0.6 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.3 | 1.2 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.3 | 1.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 | 1.5 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.3 | 0.9 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.3 | 3.8 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.3 | 0.9 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.3 | 1.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.3 | 4.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.3 | 1.2 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.3 | 0.9 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.3 | 1.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.3 | 0.9 | GO:0090076 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 4.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.3 | 1.4 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.3 | 3.0 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.3 | 1.9 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.3 | 4.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) |
| 0.3 | 1.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 1.9 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.3 | 0.8 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.3 | 2.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.3 | 2.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 0.8 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.3 | 0.8 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 1.3 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.3 | 2.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 0.5 | GO:0021586 | pons maturation(GO:0021586) |
| 0.3 | 1.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 1.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 2.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 2.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 1.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 1.7 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.2 | 3.6 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.2 | 2.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.2 | 1.4 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.2 | 2.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 1.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 0.5 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.2 | 1.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 1.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 6.0 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.4 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 3.8 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.2 | 5.9 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 2.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 3.6 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.2 | 0.4 | GO:0097017 | renal protein absorption(GO:0097017) |
| 0.2 | 2.6 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 1.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 2.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 1.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 4.7 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.2 | 3.4 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 3.4 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.2 | 3.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 7.2 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 2.5 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 1.9 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 1.0 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.2 | 1.0 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 2.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.2 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.2 | 3.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 3.5 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.2 | 0.6 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.2 | 7.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 4.4 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.2 | 0.6 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 2.5 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.2 | 1.4 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 1.7 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 | 0.6 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 0.4 | GO:0035247 | peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.2 | 0.8 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.2 | 1.9 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.2 | 0.4 | GO:1902338 | negative regulation of apoptotic process involved in morphogenesis(GO:1902338) |
| 0.2 | 0.5 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.2 | 1.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 0.5 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 2.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 1.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 5.1 | GO:0070570 | regulation of neuron projection regeneration(GO:0070570) |
| 0.2 | 0.9 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.2 | 2.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.9 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 0.7 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.2 | 5.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 1.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 5.4 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.2 | 1.8 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.2 | 1.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 2.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 0.2 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.2 | 1.5 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.2 | 1.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) male mating behavior(GO:0060179) |
| 0.2 | 0.8 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 3.2 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.2 | 2.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 0.6 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 4.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 6.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.6 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.2 | 1.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 2.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 2.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 1.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 3.4 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 0.7 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 3.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.0 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.1 | 1.9 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 1.0 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.5 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 0.7 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.3 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 2.8 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.9 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.3 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 1.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.3 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 0.1 | 1.0 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 4.6 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 1.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.4 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.9 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 1.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 14.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 3.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 2.9 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 1.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 1.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 1.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.7 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 3.1 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.1 | 1.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 8.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.4 | GO:0060770 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 1.2 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 0.8 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 1.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.9 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 1.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 4.2 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 0.5 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.4 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 1.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.4 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 2.7 | GO:2000757 | negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.1 | 1.0 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.6 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 1.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 1.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.6 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 1.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 3.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 3.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 2.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 2.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.6 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 2.4 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 1.4 | GO:1904851 | positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.1 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 1.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 2.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.6 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.1 | 2.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 1.2 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.1 | 0.2 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.2 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.1 | 1.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.7 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.3 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.4 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.8 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 2.9 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 1.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 2.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.9 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.8 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.4 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.5 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 0.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.3 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 3.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 2.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.2 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 1.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.0 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.9 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.6 | GO:0048668 | collateral sprouting(GO:0048668) |
| 0.1 | 3.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.7 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 0.7 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 7.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 0.6 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 1.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 1.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.7 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 1.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 2.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 1.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 7.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.4 | GO:1904874 | positive regulation of telomerase RNA localization to Cajal body(GO:1904874) |
| 0.1 | 1.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.1 | 1.7 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 2.8 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:1902748 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 1.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.4 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 1.3 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 0.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.8 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.0 | 1.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.7 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.6 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 2.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 1.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0099645 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) NMDA selective glutamate receptor signaling pathway(GO:0098989) neurotransmitter receptor diffusion trapping(GO:0099628) protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 1.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 1.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.3 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.0 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.1 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 1.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.8 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 2.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.9 | GO:0051497 | negative regulation of actin filament bundle assembly(GO:0032232) negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 2.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 2.5 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.3 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 1.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 3.1 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 2.1 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 1.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 2.4 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.6 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 1.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 2.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.1 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.0 | 0.8 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.3 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 1.0 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 2.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.1 | GO:0035128 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 | 3.4 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 1.0 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 1.1 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.2 | GO:1900271 | regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 1.1 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 1.0 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 1.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 2.9 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.6 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0097447 | dendritic tree(GO:0097447) |
| 1.1 | 3.4 | GO:0044753 | amphisome(GO:0044753) |
| 1.1 | 3.4 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 1.1 | 8.8 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 1.0 | 6.7 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.9 | 3.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.8 | 2.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.7 | 2.1 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.7 | 10.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.7 | 2.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.6 | 4.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.6 | 7.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.6 | 2.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.6 | 2.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.5 | 23.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.5 | 3.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.5 | 3.5 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.5 | 2.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.5 | 2.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.5 | 8.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 5.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.4 | 6.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.4 | 2.2 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.4 | 2.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.4 | 1.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.4 | 1.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 2.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.4 | 7.0 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 2.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.3 | 3.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 2.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 2.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.3 | 1.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 2.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.3 | 7.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 6.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 8.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 0.9 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.3 | 17.3 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 4.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 1.7 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 2.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 7.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 1.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 2.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.3 | 2.2 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 3.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 1.8 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 1.5 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 3.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 5.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 2.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 2.4 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 0.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 3.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 7.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.6 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 1.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 4.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 3.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 2.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 1.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 5.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 11.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 2.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 2.1 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 0.5 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 1.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 0.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 6.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.7 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.3 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 3.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 3.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 1.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 3.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 2.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 1.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 5.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 2.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 3.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.5 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 2.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.3 | GO:0060473 | cortical granule(GO:0060473) |
| 0.1 | 1.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 9.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 21.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 3.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 4.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 3.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.3 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 44.6 | GO:0098794 | postsynapse(GO:0098794) |
| 0.1 | 0.8 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 0.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 9.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 3.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 3.8 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 1.1 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 3.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.7 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 5.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 23.3 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.1 | 4.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 8.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 16.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 3.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 9.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 4.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 3.7 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 1.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 2.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 5.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 10.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 34.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.1 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 2.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 2.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.8 | 7.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.5 | 8.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 1.2 | 3.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.1 | 2.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.1 | 3.4 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.0 | 3.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.9 | 3.7 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.9 | 3.7 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.9 | 2.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.9 | 2.6 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.8 | 4.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.8 | 4.9 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.8 | 4.0 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.8 | 5.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.8 | 10.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.7 | 10.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.7 | 2.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.7 | 11.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 4.6 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.6 | 3.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.6 | 2.5 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.6 | 3.7 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.6 | 3.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.6 | 1.8 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.6 | 3.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.6 | 6.9 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.6 | 6.9 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.6 | 2.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.5 | 3.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.5 | 3.2 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.5 | 1.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.5 | 9.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 3.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.5 | 1.4 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.5 | 7.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.4 | 6.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 1.3 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.4 | 4.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 2.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.4 | 4.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 4.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 1.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.4 | 4.6 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 2.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.4 | 1.7 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.4 | 3.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 1.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 5.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 1.2 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.4 | 9.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.4 | 3.9 | GO:0070061 | fructose binding(GO:0070061) |
| 0.4 | 2.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.3 | 1.7 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.3 | 1.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.3 | 2.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 11.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 2.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.3 | 4.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.3 | 1.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.3 | 1.6 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.3 | 1.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 4.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 11.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 2.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 2.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 3.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 0.9 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.3 | 6.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 7.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 0.9 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.3 | 0.9 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.3 | 7.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 1.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 3.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 1.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.3 | 1.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.3 | 2.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.3 | 2.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 2.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 5.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 2.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 4.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.3 | 1.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.3 | 1.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 1.3 | GO:2001070 | starch binding(GO:2001070) |
| 0.3 | 1.0 | GO:0004844 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 1.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 1.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 0.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 1.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 1.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 3.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 5.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 0.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 5.6 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.2 | 1.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 2.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.5 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.2 | 3.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.6 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 1.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 2.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.6 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.2 | 6.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 3.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 1.8 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.2 | 0.6 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 1.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 5.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 4.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 3.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.8 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.2 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 1.7 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.2 | 8.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 4.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 3.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 7.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 2.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 2.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 5.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 3.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 4.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 2.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 0.8 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 0.5 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 4.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 1.4 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.2 | 1.5 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 1.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 2.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 2.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 6.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 1.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.1 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 1.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 2.0 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.9 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 2.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 11.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 4.7 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.1 | 1.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.4 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 3.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 4.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 1.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 10.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 3.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 5.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.8 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.3 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.9 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.4 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.3 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.1 | 2.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.6 | GO:0005131 | growth hormone receptor binding(GO:0005131) JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.9 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 3.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 3.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 5.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 2.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.3 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 0.6 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 9.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 2.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.8 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 1.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.9 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) lithocholic acid binding(GO:1902121) |
| 0.1 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.5 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 1.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 16.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 3.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.9 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 1.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 3.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 3.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.4 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 0.7 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.3 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.1 | 7.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 3.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.2 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 0.2 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 2.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 2.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 2.1 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.1 | 0.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 1.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 13.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 1.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 3.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 3.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 3.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.0 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 27.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 4.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.8 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 8.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.3 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.8 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 5.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 1.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 8.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 1.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 4.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 1.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 3.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 3.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 7.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 6.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 4.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 10.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 8.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 5.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 13.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 4.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 6.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 0.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 9.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 7.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 4.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 8.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 5.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 2.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 0.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 0.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 3.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 6.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 10.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 2.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 2.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 12.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 6.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 17.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 8.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.3 | 6.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 12.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 16.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 9.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 2.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 4.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 14.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 6.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 3.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.2 | 2.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 4.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 3.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 5.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 2.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 5.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 6.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 4.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 6.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 6.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 7.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 8.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 4.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 11.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 4.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 2.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 0.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 3.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.1 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.6 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |