Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
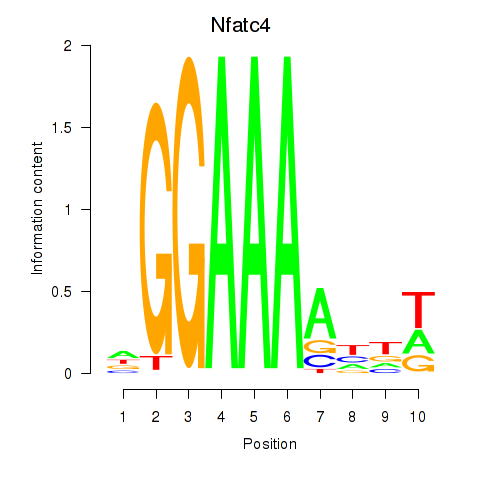
Results for Nfatc4
Z-value: 0.42
Transcription factors associated with Nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc4
|
ENSMUSG00000023411.13 | Nfatc4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc4 | mm39_v1_chr14_+_56062252_56062348 | 0.13 | 2.9e-01 | Click! |
Activity profile of Nfatc4 motif
Sorted Z-values of Nfatc4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_33071964 | 2.81 |
ENSMUST00000219211.2
|
Trdn
|
triadin |
| chr9_+_77543776 | 1.90 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
| chr11_-_47270201 | 1.50 |
ENSMUST00000077221.6
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr13_+_83652352 | 1.41 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr5_-_112542671 | 1.36 |
ENSMUST00000196256.2
|
Asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chrX_+_7504913 | 1.36 |
ENSMUST00000128890.2
|
Syp
|
synaptophysin |
| chr4_+_48049080 | 1.29 |
ENSMUST00000153369.2
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr14_+_70793340 | 1.28 |
ENSMUST00000163060.2
|
Hr
|
lysine demethylase and nuclear receptor corepressor |
| chr1_-_150341911 | 1.18 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr16_+_38182569 | 1.16 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr12_-_84497718 | 1.16 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr13_+_83652150 | 1.14 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr2_-_73490719 | 1.06 |
ENSMUST00000154258.8
|
Chn1
|
chimerin 1 |
| chr13_+_83652280 | 1.04 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr19_-_8816530 | 0.97 |
ENSMUST00000096259.6
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr2_+_55325931 | 0.93 |
ENSMUST00000067101.10
|
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr3_-_20329823 | 0.86 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chrX_+_162923474 | 0.86 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr9_+_113641615 | 0.83 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr19_-_4993060 | 0.82 |
ENSMUST00000133504.2
ENSMUST00000133254.2 ENSMUST00000120475.8 ENSMUST00000025834.15 |
Peli3
|
pellino 3 |
| chr12_+_84498196 | 0.79 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr2_+_61876806 | 0.79 |
ENSMUST00000102735.10
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr13_-_53135064 | 0.66 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr19_+_8828132 | 0.66 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr5_+_117501557 | 0.63 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr17_+_48037758 | 0.63 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr10_-_44334683 | 0.62 |
ENSMUST00000105490.3
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chrX_-_23231245 | 0.62 |
ENSMUST00000115313.8
|
Klhl13
|
kelch-like 13 |
| chr12_-_86125793 | 0.61 |
ENSMUST00000003687.8
|
Tgfb3
|
transforming growth factor, beta 3 |
| chr6_-_21851827 | 0.60 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr14_+_120513106 | 0.60 |
ENSMUST00000227012.2
ENSMUST00000167459.3 |
Mbnl2
|
muscleblind like splicing factor 2 |
| chr3_+_84832783 | 0.59 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr7_-_79570342 | 0.54 |
ENSMUST00000075657.8
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr1_-_169938298 | 0.52 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr18_+_73996743 | 0.50 |
ENSMUST00000134847.2
|
Mro
|
maestro |
| chr1_-_169938060 | 0.48 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr10_-_12689345 | 0.47 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr1_+_34044940 | 0.47 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr10_+_21869776 | 0.46 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chrX_-_166907286 | 0.45 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr2_+_174171799 | 0.45 |
ENSMUST00000109085.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr6_+_21986445 | 0.44 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr2_+_174171860 | 0.43 |
ENSMUST00000109087.8
ENSMUST00000109084.8 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr14_+_120513076 | 0.43 |
ENSMUST00000088419.13
|
Mbnl2
|
muscleblind like splicing factor 2 |
| chr10_-_44334711 | 0.41 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr2_+_124978518 | 0.41 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr14_-_79718890 | 0.41 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr8_-_41494890 | 0.39 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr1_-_168259710 | 0.37 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr2_+_124978612 | 0.36 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr2_+_3115250 | 0.36 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr7_+_91321500 | 0.32 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr9_+_114159203 | 0.29 |
ENSMUST00000066460.5
|
Bcl2a1c
|
B cell leukemia/lymphoma 2 related protein A1c |
| chr8_-_68270936 | 0.27 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr4_-_148244299 | 0.27 |
ENSMUST00000151127.8
ENSMUST00000105705.9 |
Fbxo44
|
F-box protein 44 |
| chr1_-_126758369 | 0.26 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr3_-_96501443 | 0.26 |
ENSMUST00000145001.2
ENSMUST00000091924.10 |
Polr3gl
|
polymerase (RNA) III (DNA directed) polypeptide G like |
| chr2_-_66240408 | 0.26 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr17_-_49871291 | 0.25 |
ENSMUST00000224595.2
ENSMUST00000057610.8 |
Daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr8_-_39128662 | 0.24 |
ENSMUST00000118896.2
|
Sgcz
|
sarcoglycan zeta |
| chr3_-_151899470 | 0.24 |
ENSMUST00000050073.13
|
Dnajb4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr8_+_129085719 | 0.21 |
ENSMUST00000026917.10
|
Nrp1
|
neuropilin 1 |
| chr15_-_79816717 | 0.19 |
ENSMUST00000177044.2
ENSMUST00000109615.8 |
Cbx7
|
chromobox 7 |
| chr6_-_128599779 | 0.14 |
ENSMUST00000203275.3
|
Klrb1a
|
killer cell lectin-like receptor subfamily B member 1A |
| chr11_-_89587671 | 0.13 |
ENSMUST00000128717.9
|
Ankfn1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr9_+_107869662 | 0.13 |
ENSMUST00000177173.8
|
Cdhr4
|
cadherin-related family member 4 |
| chr6_+_134958681 | 0.13 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr18_+_42186713 | 0.12 |
ENSMUST00000072008.11
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr7_+_80764564 | 0.11 |
ENSMUST00000119083.2
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chrX_+_56144946 | 0.11 |
ENSMUST00000141936.2
|
Vgll1
|
vestigial like family member 1 |
| chr13_+_25127127 | 0.11 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr2_-_73284262 | 0.11 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr12_+_103564479 | 0.09 |
ENSMUST00000190151.2
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr15_-_79816785 | 0.09 |
ENSMUST00000089293.11
ENSMUST00000109616.9 |
Cbx7
|
chromobox 7 |
| chr7_+_80764547 | 0.06 |
ENSMUST00000026820.11
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr14_-_65187287 | 0.06 |
ENSMUST00000067843.10
ENSMUST00000176489.8 ENSMUST00000175905.8 ENSMUST00000022544.14 ENSMUST00000175744.8 ENSMUST00000176128.8 |
Hmbox1
|
homeobox containing 1 |
| chr12_+_76580386 | 0.06 |
ENSMUST00000219063.2
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr10_+_23727325 | 0.05 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr3_-_86455575 | 0.04 |
ENSMUST00000077524.4
|
Mab21l2
|
mab-21-like 2 |
| chr18_+_42186757 | 0.03 |
ENSMUST00000074679.4
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr11_-_99412084 | 0.03 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chr16_+_3656839 | 0.02 |
ENSMUST00000080917.2
|
Olfr15
|
olfactory receptor 15 |
| chr11_-_99412162 | 0.00 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.5 | 1.4 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.3 | 1.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.3 | 1.3 | GO:1900623 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 3.6 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.3 | 1.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.2 | 1.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.2 | 0.9 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.2 | 0.6 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 0.6 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 1.2 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 0.6 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 1.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.5 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 | 0.2 | GO:0097374 | cell migration involved in vasculogenesis(GO:0035441) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.5 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 1.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.5 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.2 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0010982 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.0 | 0.5 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 1.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 0.8 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.2 | 1.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.8 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 1.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 1.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.8 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 1.9 | GO:0042383 | sarcolemma(GO:0042383) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 3.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 1.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.9 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.2 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 1.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 1.2 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.4 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 0.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.9 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |