Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
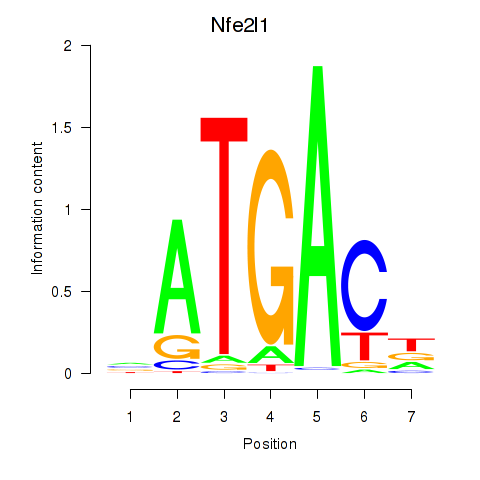
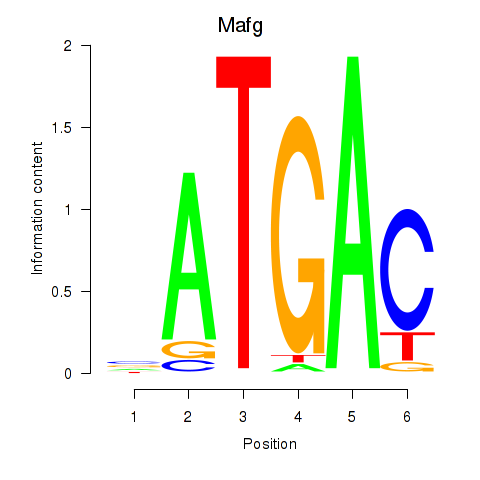
Results for Nfe2l1_Mafg
Z-value: 0.93
Transcription factors associated with Nfe2l1_Mafg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l1
|
ENSMUSG00000038615.18 | Nfe2l1 |
|
Mafg
|
ENSMUSG00000051510.14 | Mafg |
|
Mafg
|
ENSMUSG00000051510.14 | Mafg |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafg | mm39_v1_chr11_-_120524362_120524431 | -0.68 | 4.5e-11 | Click! |
| Nfe2l1 | mm39_v1_chr11_-_96714813_96714850 | -0.51 | 5.4e-06 | Click! |
Activity profile of Nfe2l1_Mafg motif
Sorted Z-values of Nfe2l1_Mafg motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2l1_Mafg
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_122129379 | 8.99 |
ENSMUST00000028656.2
|
Duoxa2
|
dual oxidase maturation factor 2 |
| chr5_+_33261563 | 7.16 |
ENSMUST00000011178.5
|
Slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr2_-_60503998 | 4.78 |
ENSMUST00000059888.15
ENSMUST00000154764.2 |
Itgb6
|
integrin beta 6 |
| chr1_-_82746169 | 4.64 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr1_+_88015524 | 4.53 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr11_+_102036356 | 4.31 |
ENSMUST00000055409.6
|
Nags
|
N-acetylglutamate synthase |
| chr1_-_66984178 | 4.23 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr17_-_41191414 | 4.21 |
ENSMUST00000068258.3
ENSMUST00000233122.2 |
9130008F23Rik
|
RIKEN cDNA 9130008F23 gene |
| chr1_-_190915441 | 3.93 |
ENSMUST00000027941.14
|
Atf3
|
activating transcription factor 3 |
| chr13_+_38335302 | 3.78 |
ENSMUST00000127906.8
|
Dsp
|
desmoplakin |
| chr17_+_44445659 | 3.66 |
ENSMUST00000239215.2
|
Clic5
|
chloride intracellular channel 5 |
| chr13_+_38335232 | 3.55 |
ENSMUST00000124830.3
|
Dsp
|
desmoplakin |
| chr17_-_48739874 | 3.53 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr17_-_35954573 | 3.53 |
ENSMUST00000095467.4
|
Mucl3
|
mucin like 3 |
| chr1_-_66984521 | 3.45 |
ENSMUST00000160100.2
|
Myl1
|
myosin, light polypeptide 1 |
| chr3_-_146302343 | 3.11 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr7_-_115637970 | 3.08 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr1_-_128256048 | 3.01 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr11_+_98555167 | 2.98 |
ENSMUST00000017348.3
|
Gsdma
|
gasdermin A |
| chr10_+_75983285 | 2.85 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr1_+_67162176 | 2.81 |
ENSMUST00000027144.8
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr10_-_13744676 | 2.72 |
ENSMUST00000019942.6
ENSMUST00000162610.8 |
Aig1
|
androgen-induced 1 |
| chr10_-_13744523 | 2.47 |
ENSMUST00000105534.10
|
Aig1
|
androgen-induced 1 |
| chr9_+_35122451 | 2.42 |
ENSMUST00000034541.12
|
Srpr
|
signal recognition particle receptor ('docking protein') |
| chr12_-_103304573 | 2.42 |
ENSMUST00000149431.2
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr3_+_3699205 | 2.28 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr11_-_83421333 | 2.21 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr2_-_76698725 | 2.16 |
ENSMUST00000149616.8
ENSMUST00000152185.8 ENSMUST00000130915.8 ENSMUST00000155365.8 ENSMUST00000128071.8 |
Ttn
|
titin |
| chr3_-_144514386 | 2.09 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr5_-_46013838 | 2.04 |
ENSMUST00000087164.10
ENSMUST00000121573.8 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr11_-_51891259 | 2.02 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr5_-_66309244 | 1.99 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr7_+_144391786 | 1.99 |
ENSMUST00000155320.8
|
Fgf3
|
fibroblast growth factor 3 |
| chr13_-_3854307 | 1.95 |
ENSMUST00000077698.5
|
Calml3
|
calmodulin-like 3 |
| chr8_-_36293588 | 1.87 |
ENSMUST00000060128.7
|
Cldn23
|
claudin 23 |
| chr15_-_101833160 | 1.78 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr5_-_51711237 | 1.71 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr3_-_85648696 | 1.70 |
ENSMUST00000094148.6
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr2_-_52225146 | 1.67 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr5_-_73787646 | 1.67 |
ENSMUST00000152215.3
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr15_-_41733099 | 1.64 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein |
| chr5_-_51711204 | 1.64 |
ENSMUST00000196968.5
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr6_+_142244145 | 1.63 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr16_-_88360037 | 1.62 |
ENSMUST00000049697.5
|
Cldn8
|
claudin 8 |
| chr6_-_24528012 | 1.55 |
ENSMUST00000023851.9
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr4_-_43821433 | 1.54 |
ENSMUST00000079465.5
|
Olfr156
|
olfactory receptor 156 |
| chr2_-_52225763 | 1.53 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr3_-_91990439 | 1.52 |
ENSMUST00000058150.8
|
Lor
|
loricrin |
| chrX_+_56088434 | 1.45 |
ENSMUST00000033464.4
|
Brs3
|
bombesin-like receptor 3 |
| chr10_-_75946790 | 1.43 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr6_-_86646118 | 1.43 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr2_-_155676765 | 1.41 |
ENSMUST00000029143.7
ENSMUST00000239423.2 |
Fam83c
|
family with sequence similarity 83, member C |
| chr6_-_70435020 | 1.40 |
ENSMUST00000198184.2
|
Igkv6-13
|
immunoglobulin kappa variable 6-13 |
| chr1_-_20854490 | 1.39 |
ENSMUST00000039046.10
|
Il17f
|
interleukin 17F |
| chr4_-_131776368 | 1.38 |
ENSMUST00000105981.9
ENSMUST00000084253.10 ENSMUST00000141291.3 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr19_-_36097233 | 1.35 |
ENSMUST00000025718.10
|
Ankrd1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr15_+_102829538 | 1.33 |
ENSMUST00000001700.7
|
Hoxc13
|
homeobox C13 |
| chr9_-_122942196 | 1.32 |
ENSMUST00000238520.2
ENSMUST00000147563.9 |
Zdhhc3
|
zinc finger, DHHC domain containing 3 |
| chr1_-_65090278 | 1.30 |
ENSMUST00000161960.2
ENSMUST00000087359.6 |
Cryge
|
crystallin, gamma E |
| chr14_+_8293646 | 1.29 |
ENSMUST00000035250.6
|
Olfr720
|
olfactory receptor 720 |
| chr11_-_51891575 | 1.28 |
ENSMUST00000109086.8
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr19_+_53781721 | 1.27 |
ENSMUST00000162910.2
|
Rbm20
|
RNA binding motif protein 20 |
| chr2_-_150531280 | 1.27 |
ENSMUST00000046095.10
|
Vsx1
|
visual system homeobox 1 |
| chr5_-_116221293 | 1.25 |
ENSMUST00000111997.3
ENSMUST00000239438.2 |
Tmem233
|
transmembrane protein 233 |
| chr6_+_36364990 | 1.23 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr9_+_37903788 | 1.23 |
ENSMUST00000074611.3
|
Olfr881
|
olfactory receptor 881 |
| chr11_+_116324913 | 1.21 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2 |
| chr19_-_12897671 | 1.21 |
ENSMUST00000054737.3
|
Olfr1448
|
olfactory receptor 1448 |
| chr17_+_87590308 | 1.20 |
ENSMUST00000041110.12
ENSMUST00000125875.8 |
Ttc7
|
tetratricopeptide repeat domain 7 |
| chr2_-_63928339 | 1.18 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr15_-_101602734 | 1.18 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A |
| chr3_-_79474989 | 1.16 |
ENSMUST00000076136.7
|
Fnip2
|
folliculin interacting protein 2 |
| chr7_+_118232952 | 1.16 |
ENSMUST00000057320.8
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chr4_+_33081505 | 1.15 |
ENSMUST00000147889.2
|
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr11_+_98517170 | 1.13 |
ENSMUST00000073295.3
ENSMUST00000107508.9 |
Gsdma3
|
gasdermin A3 |
| chr1_+_87998487 | 1.11 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr10_+_23825456 | 1.09 |
ENSMUST00000045152.6
|
Taar3
|
trace amine-associated receptor 3 |
| chr19_-_11261177 | 1.09 |
ENSMUST00000186937.7
ENSMUST00000067673.13 |
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr5_-_113285852 | 1.09 |
ENSMUST00000212276.2
|
2900026A02Rik
|
RIKEN cDNA 2900026A02 gene |
| chr9_+_37766116 | 1.05 |
ENSMUST00000086063.4
|
Olfr877
|
olfactory receptor 877 |
| chr10_+_51898598 | 1.05 |
ENSMUST00000163017.10
|
Vgll2
|
vestigial like family member 2 |
| chr12_-_87691390 | 1.04 |
ENSMUST00000220528.2
|
Ubl5b
|
ubiquitin-like 5B |
| chr11_-_65160767 | 1.03 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr1_-_36312482 | 1.03 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr6_-_70412460 | 1.02 |
ENSMUST00000103394.2
|
Igkv6-14
|
immunoglobulin kappa variable 6-14 |
| chr11_-_65160810 | 1.01 |
ENSMUST00000108695.9
|
Myocd
|
myocardin |
| chr17_-_46956920 | 1.01 |
ENSMUST00000233974.2
|
Klc4
|
kinesin light chain 4 |
| chr7_+_112278520 | 1.01 |
ENSMUST00000084705.13
ENSMUST00000239442.2 ENSMUST00000239404.2 ENSMUST00000059768.18 |
Tead1
|
TEA domain family member 1 |
| chr6_+_40468167 | 1.00 |
ENSMUST00000064932.6
|
Tas2r137
|
taste receptor, type 2, member 137 |
| chr8_-_85740220 | 0.98 |
ENSMUST00000209322.2
|
Best2
|
bestrophin 2 |
| chr15_-_101801351 | 0.98 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr4_-_119272667 | 0.95 |
ENSMUST00000238609.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr15_+_98335351 | 0.95 |
ENSMUST00000075851.4
|
Olfr282
|
olfactory receptor 282 |
| chr14_-_51308899 | 0.95 |
ENSMUST00000048478.6
|
Olfr750
|
olfactory receptor 750 |
| chr3_-_144511566 | 0.95 |
ENSMUST00000199029.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr10_-_23985432 | 0.94 |
ENSMUST00000041180.7
|
Taar9
|
trace amine-associated receptor 9 |
| chr5_-_139812162 | 0.94 |
ENSMUST00000182839.2
|
Gm26938
|
predicted gene, 26938 |
| chr19_+_12245527 | 0.92 |
ENSMUST00000073507.3
|
Olfr235
|
olfactory receptor 235 |
| chr7_+_104718676 | 0.90 |
ENSMUST00000098160.2
|
Olfr678
|
olfactory receptor 678 |
| chr1_-_133849131 | 0.89 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr11_-_86574586 | 0.88 |
ENSMUST00000018315.10
|
Vmp1
|
vacuole membrane protein 1 |
| chr14_-_118289557 | 0.86 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr7_+_112278534 | 0.84 |
ENSMUST00000106638.10
|
Tead1
|
TEA domain family member 1 |
| chr12_-_81014849 | 0.83 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chrX_-_161671421 | 0.82 |
ENSMUST00000033723.4
|
Syap1
|
synapse associated protein 1 |
| chr10_+_93871863 | 0.82 |
ENSMUST00000020208.5
|
Fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr12_-_81014755 | 0.81 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr9_-_22041894 | 0.80 |
ENSMUST00000115315.3
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr9_+_53212871 | 0.79 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr9_+_108765701 | 0.79 |
ENSMUST00000026743.14
ENSMUST00000194047.3 |
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr6_-_83098255 | 0.77 |
ENSMUST00000205023.2
ENSMUST00000146328.4 ENSMUST00000151393.7 ENSMUST00000032111.11 ENSMUST00000113936.10 |
Wbp1
|
WW domain binding protein 1 |
| chr3_-_59102517 | 0.77 |
ENSMUST00000200095.2
|
Gpr87
|
G protein-coupled receptor 87 |
| chr13_-_18556626 | 0.77 |
ENSMUST00000139064.10
ENSMUST00000175703.9 |
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr1_+_173986288 | 0.77 |
ENSMUST00000068403.4
|
Olfr420
|
olfactory receptor 420 |
| chr17_-_58049214 | 0.77 |
ENSMUST00000178907.2
|
Gm21834
|
predicted gene, 21834 |
| chr11_-_50216426 | 0.76 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr7_-_104777612 | 0.76 |
ENSMUST00000214399.4
|
Olfr682-ps1
|
olfactory receptor 682, pseudogene 1 |
| chr19_+_12186932 | 0.76 |
ENSMUST00000072316.5
|
Olfr1431
|
olfactory receptor 1431 |
| chr11_+_72498029 | 0.76 |
ENSMUST00000021148.13
ENSMUST00000138247.8 |
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chrX_+_70707271 | 0.75 |
ENSMUST00000070449.6
|
Gpr50
|
G-protein-coupled receptor 50 |
| chr10_+_23836392 | 0.75 |
ENSMUST00000092660.2
|
Taar4
|
trace amine-associated receptor 4 |
| chrM_+_7006 | 0.75 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr12_+_69418886 | 0.74 |
ENSMUST00000050063.9
|
Arf6
|
ADP-ribosylation factor 6 |
| chr6_+_17463925 | 0.74 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr14_-_52728503 | 0.73 |
ENSMUST00000073571.6
|
Olfr1507
|
olfactory receptor 1507 |
| chr18_-_9282754 | 0.73 |
ENSMUST00000041007.4
|
Gjd4
|
gap junction protein, delta 4 |
| chr8_+_71917554 | 0.71 |
ENSMUST00000048914.8
|
Mrpl34
|
mitochondrial ribosomal protein L34 |
| chrX_-_48877080 | 0.71 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr2_-_76700830 | 0.71 |
ENSMUST00000138542.2
|
Ttn
|
titin |
| chr5_-_135378896 | 0.69 |
ENSMUST00000201534.2
ENSMUST00000044972.11 |
Fkbp6
|
FK506 binding protein 6 |
| chr17_-_35077089 | 0.68 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr3_-_79475078 | 0.68 |
ENSMUST00000133154.7
|
Fnip2
|
folliculin interacting protein 2 |
| chr1_+_173964312 | 0.67 |
ENSMUST00000053941.4
|
Olfr424
|
olfactory receptor 424 |
| chr17_-_34822649 | 0.66 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr19_-_12206908 | 0.66 |
ENSMUST00000180978.3
|
Olfr1432
|
olfactory receptor 1432 |
| chr5_-_107873883 | 0.65 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr7_+_139902515 | 0.65 |
ENSMUST00000078103.3
|
Olfr525
|
olfactory receptor 525 |
| chr10_+_59715378 | 0.64 |
ENSMUST00000147914.8
ENSMUST00000146590.8 |
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr8_-_37420293 | 0.63 |
ENSMUST00000179501.2
|
Dlc1
|
deleted in liver cancer 1 |
| chr10_-_62723103 | 0.63 |
ENSMUST00000218438.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr9_+_39933648 | 0.63 |
ENSMUST00000059859.5
|
Olfr981
|
olfactory receptor 981 |
| chr11_+_100978103 | 0.63 |
ENSMUST00000107302.8
ENSMUST00000107303.10 ENSMUST00000017945.15 ENSMUST00000149597.2 |
Mlx
|
MAX-like protein X |
| chr10_+_59715439 | 0.62 |
ENSMUST00000142819.8
ENSMUST00000020309.7 |
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr5_+_31070739 | 0.62 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr18_+_82929451 | 0.61 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr17_-_42922286 | 0.60 |
ENSMUST00000068355.8
|
Opn5
|
opsin 5 |
| chr7_+_108312527 | 0.59 |
ENSMUST00000209620.4
ENSMUST00000074730.4 |
Olfr512
|
olfactory receptor 512 |
| chrX_+_45417884 | 0.58 |
ENSMUST00000059466.3
|
Actrt1
|
actin-related protein T1 |
| chr9_-_35122261 | 0.57 |
ENSMUST00000043805.15
ENSMUST00000142595.8 ENSMUST00000127996.8 |
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr12_+_84408803 | 0.57 |
ENSMUST00000110278.8
ENSMUST00000145522.2 |
Coq6
|
coenzyme Q6 monooxygenase |
| chr10_-_62723238 | 0.56 |
ENSMUST00000228901.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr5_+_100054110 | 0.56 |
ENSMUST00000198837.3
|
Vamp9
|
vesicle-associated membrane protein 9 |
| chr5_-_135378729 | 0.56 |
ENSMUST00000201784.4
ENSMUST00000201791.4 |
Fkbp6
|
FK506 binding protein 6 |
| chr10_+_34265969 | 0.55 |
ENSMUST00000105511.2
|
Col10a1
|
collagen, type X, alpha 1 |
| chr17_-_64170786 | 0.55 |
ENSMUST00000050753.4
|
A930002H24Rik
|
RIKEN cDNA A930002H24 gene |
| chr1_+_20801127 | 0.55 |
ENSMUST00000027061.5
|
Il17a
|
interleukin 17A |
| chr3_+_135144287 | 0.54 |
ENSMUST00000196591.5
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_+_140190081 | 0.54 |
ENSMUST00000072655.3
|
Olfr46
|
olfactory receptor 46 |
| chr12_+_84408742 | 0.54 |
ENSMUST00000021661.13
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr19_+_13211616 | 0.54 |
ENSMUST00000064102.2
|
Olfr1463
|
olfactory receptor 1463 |
| chr15_-_101848689 | 0.53 |
ENSMUST00000023799.8
|
Krt79
|
keratin 79 |
| chr3_+_135144304 | 0.53 |
ENSMUST00000198685.5
ENSMUST00000197859.5 |
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_58785722 | 0.53 |
ENSMUST00000059608.5
|
Olfr267
|
olfactory receptor 267 |
| chr11_-_58611920 | 0.53 |
ENSMUST00000189911.3
|
Olfr318
|
olfactory receptor 318 |
| chr2_+_85551751 | 0.52 |
ENSMUST00000055517.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr5_+_122845607 | 0.52 |
ENSMUST00000031429.14
|
P2rx4
|
purinergic receptor P2X, ligand-gated ion channel 4 |
| chr16_+_35803794 | 0.51 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr2_-_89774457 | 0.48 |
ENSMUST00000090695.3
|
Olfr1259
|
olfactory receptor 1259 |
| chr2_-_89156522 | 0.48 |
ENSMUST00000099785.2
|
Olfr1232
|
olfactory receptor 1232 |
| chr4_+_52825399 | 0.48 |
ENSMUST00000095085.2
|
Olfr275
|
olfactory receptor 275 |
| chr1_+_65965670 | 0.48 |
ENSMUST00000027082.11
ENSMUST00000114027.3 |
Crygf
|
crystallin, gamma F |
| chr13_+_111391544 | 0.47 |
ENSMUST00000054716.4
|
Actbl2
|
actin, beta-like 2 |
| chr19_+_12824046 | 0.47 |
ENSMUST00000189517.2
|
Gm5244
|
predicted pseudogene 5244 |
| chr1_+_87983099 | 0.46 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr10_-_95251327 | 0.46 |
ENSMUST00000172070.8
ENSMUST00000150432.8 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr17_-_34250616 | 0.46 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr11_+_60428788 | 0.45 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr9_+_59485475 | 0.45 |
ENSMUST00000118549.8
ENSMUST00000034840.10 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr6_-_52185674 | 0.45 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr10_+_23903120 | 0.45 |
ENSMUST00000092657.2
|
Taar7d
|
trace amine-associated receptor 7D |
| chr2_-_88942854 | 0.44 |
ENSMUST00000099795.2
|
Olfr1221
|
olfactory receptor 1221 |
| chr1_+_92516054 | 0.44 |
ENSMUST00000062964.5
|
Olfr1412
|
olfactory receptor 1412 |
| chr8_+_40964818 | 0.43 |
ENSMUST00000098817.4
|
Vps37a
|
vacuolar protein sorting 37A |
| chr7_+_104236926 | 0.43 |
ENSMUST00000098173.5
ENSMUST00000210457.2 |
Olfr654
|
olfactory receptor 654 |
| chr15_+_92495007 | 0.43 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr2_+_89711138 | 0.42 |
ENSMUST00000060795.3
|
Olfr1257
|
olfactory receptor 1257 |
| chr17_+_34251041 | 0.42 |
ENSMUST00000173354.9
|
Rxrb
|
retinoid X receptor beta |
| chr5_+_45611093 | 0.42 |
ENSMUST00000053250.5
|
Clrn2
|
clarin 2 |
| chr17_+_37535982 | 0.42 |
ENSMUST00000078209.4
|
Olfr96
|
olfactory receptor 96 |
| chr6_+_43143944 | 0.41 |
ENSMUST00000060243.5
|
Olfr437
|
olfactory receptor 437 |
| chr2_-_85269581 | 0.41 |
ENSMUST00000099924.3
|
Olfr995
|
olfactory receptor 995 |
| chr16_+_3410262 | 0.41 |
ENSMUST00000061541.2
|
Olfr161
|
olfactory receptor 161 |
| chr1_+_31215482 | 0.41 |
ENSMUST00000062560.14
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr14_-_31362909 | 0.41 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr3_+_135144202 | 0.41 |
ENSMUST00000166033.6
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr16_+_58969192 | 0.40 |
ENSMUST00000075381.3
|
Olfr195
|
olfactory receptor 195 |
| chr14_+_54433524 | 0.40 |
ENSMUST00000103718.2
|
Traj23
|
T cell receptor alpha joining 23 |
| chr1_+_127701901 | 0.40 |
ENSMUST00000112570.2
ENSMUST00000027587.15 |
Ccnt2
|
cyclin T2 |
| chr11_-_120441952 | 0.40 |
ENSMUST00000026121.3
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr11_-_116743898 | 0.40 |
ENSMUST00000190993.7
ENSMUST00000092404.13 |
Srsf2
|
serine and arginine-rich splicing factor 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 1.1 | 9.0 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 1.1 | 3.3 | GO:1904635 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 1.1 | 3.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 1.0 | 3.9 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.9 | 7.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.8 | 7.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.7 | 2.0 | GO:2000724 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.6 | 3.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.6 | 2.9 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.6 | 1.1 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.5 | 6.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 2.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.5 | 4.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.4 | 5.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 4.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.3 | 1.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.3 | 0.9 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.3 | 3.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.3 | 1.4 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.2 | 1.6 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 3.7 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.2 | 0.7 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.2 | 2.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.7 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.7 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 3.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.4 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.7 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 2.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.3 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.1 | 0.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.3 | GO:0009629 | response to gravity(GO:0009629) |
| 0.1 | 1.3 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.7 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.9 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.3 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 0.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 1.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.8 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.5 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 0.3 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.1 | 1.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.4 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 0.5 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 1.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 2.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.2 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 1.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 4.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.3 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 | 0.2 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 1.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 1.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 2.1 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.8 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.0 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 1.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 3.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 1.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.7 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 1.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.5 | GO:2001028 | positive regulation of endothelial cell chemotaxis(GO:2001028) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.3 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.8 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 27.7 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 2.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.2 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 3.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 1.2 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.3 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.3 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.3 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 2.3 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 1.1 | 3.3 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 1.0 | 4.8 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.5 | 1.9 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.4 | 3.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 2.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.3 | 2.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 7.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.3 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.1 | 2.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 4.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 7.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 5.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.2 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 0.8 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 7.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 3.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.7 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 3.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 1.4 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 10.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 11.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.9 | 2.8 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.7 | 2.2 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.7 | 4.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.6 | 3.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.5 | 3.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.5 | 1.5 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.4 | 2.6 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.4 | 3.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 3.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 1.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 1.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.3 | 1.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 1.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 2.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 0.6 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.2 | 1.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.7 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 6.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.4 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.1 | 1.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 6.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 3.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 3.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.9 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.5 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 8.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.4 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 1.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 5.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 1.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 2.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 2.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 1.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 8.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 2.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 4.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 27.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 9.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 3.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 5.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 7.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 10.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 4.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 3.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 3.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 8.0 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 4.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 5.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |