Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
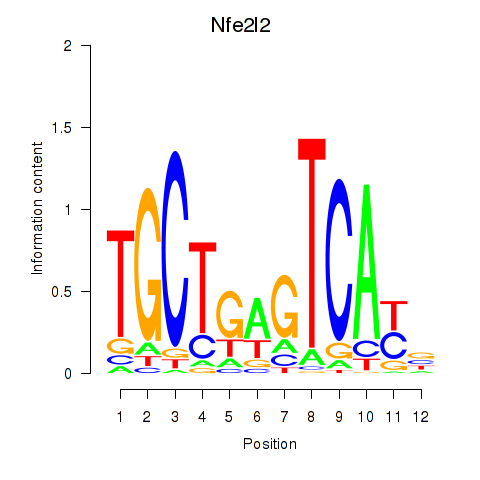
Results for Nfe2l2
Z-value: 0.98
Transcription factors associated with Nfe2l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l2
|
ENSMUSG00000015839.7 | Nfe2l2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2l2 | mm39_v1_chr2_-_75534985_75535023 | 0.14 | 2.3e-01 | Click! |
Activity profile of Nfe2l2 motif
Sorted Z-values of Nfe2l2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2l2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_44984681 | 8.10 |
ENSMUST00000085351.7
|
Hrc
|
histidine rich calcium binding protein |
| chr7_+_44984723 | 7.80 |
ENSMUST00000211327.2
|
Hrc
|
histidine rich calcium binding protein |
| chr5_+_31274046 | 6.99 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr2_-_164621641 | 6.92 |
ENSMUST00000103095.5
|
Tnnc2
|
troponin C2, fast |
| chr10_+_17672004 | 6.52 |
ENSMUST00000037964.7
|
Txlnb
|
taxilin beta |
| chr1_-_172047282 | 6.39 |
ENSMUST00000170700.2
ENSMUST00000003554.11 |
Casq1
|
calsequestrin 1 |
| chr12_-_76842263 | 6.13 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr15_-_82291372 | 5.83 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr5_+_31274064 | 5.62 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr18_-_64794338 | 5.45 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr13_-_42001075 | 4.80 |
ENSMUST00000179758.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr19_-_4092218 | 4.77 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr6_+_41928559 | 4.66 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr7_+_18817767 | 4.62 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr14_+_28740162 | 4.61 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr3_+_146302832 | 4.47 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr7_+_127399789 | 4.39 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr13_-_42001102 | 4.34 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr19_-_4087940 | 4.19 |
ENSMUST00000237893.2
ENSMUST00000169613.4 |
Gstp1
|
glutathione S-transferase, pi 1 |
| chr16_-_18904240 | 4.18 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr1_-_120001752 | 4.09 |
ENSMUST00000056089.8
|
Tmem37
|
transmembrane protein 37 |
| chr19_-_4927910 | 4.09 |
ENSMUST00000006626.5
|
Actn3
|
actinin alpha 3 |
| chr13_-_42000958 | 4.07 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr19_-_4087907 | 4.06 |
ENSMUST00000237982.2
|
Gstp1
|
glutathione S-transferase, pi 1 |
| chr1_-_162641495 | 4.01 |
ENSMUST00000144916.8
ENSMUST00000140274.2 |
Fmo4
|
flavin containing monooxygenase 4 |
| chr4_+_128999325 | 3.97 |
ENSMUST00000106064.10
ENSMUST00000030575.15 ENSMUST00000030577.11 |
Tmem54
|
transmembrane protein 54 |
| chr4_-_138484889 | 3.83 |
ENSMUST00000030526.7
|
Pla2g2f
|
phospholipase A2, group IIF |
| chr9_-_106353792 | 3.82 |
ENSMUST00000214682.2
ENSMUST00000112479.9 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr12_-_103925197 | 3.80 |
ENSMUST00000122229.8
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr7_+_127399776 | 3.70 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chrX_-_96420756 | 3.54 |
ENSMUST00000113832.2
ENSMUST00000037353.10 |
Eda2r
|
ectodysplasin A2 receptor |
| chr8_+_120163857 | 3.45 |
ENSMUST00000152420.8
ENSMUST00000212112.2 ENSMUST00000098365.4 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr16_+_22738987 | 3.44 |
ENSMUST00000023587.12
|
Fetub
|
fetuin beta |
| chr1_+_58069090 | 3.32 |
ENSMUST00000001027.7
|
Aox1
|
aldehyde oxidase 1 |
| chr9_-_106353571 | 3.29 |
ENSMUST00000123555.8
ENSMUST00000125850.2 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr4_+_116543045 | 3.29 |
ENSMUST00000129315.8
ENSMUST00000106470.8 |
Prdx1
|
peroxiredoxin 1 |
| chr3_-_116762617 | 3.29 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr17_-_28569574 | 3.24 |
ENSMUST00000114799.8
ENSMUST00000219703.3 |
Tead3
|
TEA domain family member 3 |
| chr13_-_64460491 | 3.14 |
ENSMUST00000222570.2
ENSMUST00000220895.2 |
Prxl2c
|
peroxiredoxin like 2C |
| chr18_+_4921663 | 3.06 |
ENSMUST00000143254.8
|
Svil
|
supervillin |
| chr7_+_127400016 | 2.93 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_+_11710101 | 2.91 |
ENSMUST00000138349.8
ENSMUST00000135341.8 ENSMUST00000128156.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr13_-_64460382 | 2.86 |
ENSMUST00000021938.11
ENSMUST00000221118.2 ENSMUST00000221350.2 |
Prxl2c
|
peroxiredoxin like 2C |
| chr16_+_22739191 | 2.83 |
ENSMUST00000116625.10
|
Fetub
|
fetuin beta |
| chr4_+_116542741 | 2.80 |
ENSMUST00000135573.8
ENSMUST00000151129.8 |
Prdx1
|
peroxiredoxin 1 |
| chr9_-_65330231 | 2.79 |
ENSMUST00000065894.7
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr2_-_160155536 | 2.75 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chr19_-_32080496 | 2.73 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr7_-_126398165 | 2.71 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_+_127399848 | 2.62 |
ENSMUST00000139068.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr3_-_116762476 | 2.61 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr7_-_30810422 | 2.58 |
ENSMUST00000039435.15
|
Hpn
|
hepsin |
| chr19_-_4109446 | 2.55 |
ENSMUST00000189808.7
|
Gstp3
|
glutathione S-transferase pi 3 |
| chr16_+_4825170 | 2.51 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chr10_+_75404464 | 2.46 |
ENSMUST00000145928.8
ENSMUST00000131565.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr10_+_75404482 | 2.42 |
ENSMUST00000134503.8
ENSMUST00000125770.8 ENSMUST00000128886.8 ENSMUST00000151212.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr2_+_11710246 | 2.42 |
ENSMUST00000148748.8
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr19_-_4889284 | 2.41 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr2_+_11710523 | 2.41 |
ENSMUST00000138856.2
ENSMUST00000078834.12 ENSMUST00000114834.10 ENSMUST00000114833.10 ENSMUST00000114831.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr16_+_17379749 | 2.41 |
ENSMUST00000171002.10
ENSMUST00000023441.11 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr15_-_76501525 | 2.31 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr16_+_4825146 | 2.31 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr14_-_21798678 | 2.29 |
ENSMUST00000075040.9
|
Dusp13
|
dual specificity phosphatase 13 |
| chr9_-_106353303 | 2.27 |
ENSMUST00000156426.8
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr19_-_4889314 | 2.24 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr14_+_4230569 | 2.22 |
ENSMUST00000090543.6
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr11_-_69696428 | 2.19 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr14_-_30637344 | 2.17 |
ENSMUST00000226547.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr9_-_121745354 | 2.15 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr18_+_20380397 | 2.12 |
ENSMUST00000054128.7
|
Dsg1c
|
desmoglein 1 gamma |
| chr6_+_17463925 | 2.12 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr12_-_113649535 | 2.12 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr17_-_24863956 | 2.05 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chrX_+_72719098 | 2.04 |
ENSMUST00000171398.2
|
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr7_+_45289391 | 2.03 |
ENSMUST00000148532.4
|
Mamstr
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr19_-_43928583 | 2.03 |
ENSMUST00000212396.2
|
Dnmbp
|
dynamin binding protein |
| chr13_-_23894828 | 2.01 |
ENSMUST00000091706.14
|
Hfe
|
homeostatic iron regulator |
| chr12_+_16860931 | 2.00 |
ENSMUST00000020908.9
|
E2f6
|
E2F transcription factor 6 |
| chr7_-_126398343 | 1.97 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr5_-_108823435 | 1.94 |
ENSMUST00000051757.14
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chrX_+_163052367 | 1.93 |
ENSMUST00000145412.8
ENSMUST00000033749.9 |
Pir
|
pirin |
| chr5_-_147831610 | 1.93 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr18_+_12637217 | 1.92 |
ENSMUST00000188815.2
|
Lama3
|
laminin, alpha 3 |
| chr9_+_121232480 | 1.89 |
ENSMUST00000210351.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chr1_+_157353696 | 1.86 |
ENSMUST00000111700.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr12_-_113552322 | 1.80 |
ENSMUST00000103443.3
|
Ighv2-2
|
immunoglobulin heavy variable 2-2 |
| chr14_+_4230658 | 1.78 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr6_-_68713748 | 1.77 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr18_-_35087355 | 1.72 |
ENSMUST00000025217.11
|
Hspa9
|
heat shock protein 9 |
| chr2_+_25285878 | 1.72 |
ENSMUST00000028328.3
|
Entpd2
|
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr11_+_60428788 | 1.68 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr1_+_33947250 | 1.68 |
ENSMUST00000183034.5
|
Dst
|
dystonin |
| chr1_-_121260298 | 1.65 |
ENSMUST00000071064.13
|
Insig2
|
insulin induced gene 2 |
| chr1_-_121260274 | 1.64 |
ENSMUST00000161068.2
|
Insig2
|
insulin induced gene 2 |
| chrX_-_51702813 | 1.60 |
ENSMUST00000114857.2
|
Gpc3
|
glypican 3 |
| chr12_-_17050332 | 1.56 |
ENSMUST00000134938.8
ENSMUST00000137960.2 |
Pqlc3
|
PQ loop repeat containing |
| chr16_+_90017634 | 1.55 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr18_-_61344644 | 1.53 |
ENSMUST00000146409.8
|
Slc26a2
|
solute carrier family 26 (sulfate transporter), member 2 |
| chr9_+_108566513 | 1.53 |
ENSMUST00000192344.2
|
Prkar2a
|
protein kinase, cAMP dependent regulatory, type II alpha |
| chr12_-_113617344 | 1.49 |
ENSMUST00000103447.2
|
Ighv2-4
|
immunoglobulin heavy variable V2-4 |
| chr11_+_61575245 | 1.49 |
ENSMUST00000093019.6
|
Fam83g
|
family with sequence similarity 83, member G |
| chr4_-_57956411 | 1.48 |
ENSMUST00000030051.6
|
Txn1
|
thioredoxin 1 |
| chr15_+_54434576 | 1.48 |
ENSMUST00000025356.4
|
Mal2
|
mal, T cell differentiation protein 2 |
| chr15_-_75886166 | 1.44 |
ENSMUST00000060807.12
|
Fam83h
|
family with sequence similarity 83, member H |
| chr6_+_17306334 | 1.43 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chrX_-_51702790 | 1.43 |
ENSMUST00000069360.14
|
Gpc3
|
glypican 3 |
| chr13_-_12476313 | 1.40 |
ENSMUST00000143693.8
ENSMUST00000144283.2 ENSMUST00000099820.10 ENSMUST00000135166.8 |
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr14_-_21798694 | 1.39 |
ENSMUST00000183943.2
|
Dusp13
|
dual specificity phosphatase 13 |
| chr2_+_152360167 | 1.37 |
ENSMUST00000121912.2
|
Defb28
|
defensin beta 28 |
| chr9_-_44162762 | 1.35 |
ENSMUST00000034618.6
|
Pdzd3
|
PDZ domain containing 3 |
| chr4_+_117109148 | 1.35 |
ENSMUST00000062824.12
|
Tmem53
|
transmembrane protein 53 |
| chr3_-_144514386 | 1.33 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr14_-_20529997 | 1.33 |
ENSMUST00000225132.2
|
Anxa7
|
annexin A7 |
| chr6_+_48818567 | 1.32 |
ENSMUST00000203639.3
|
Tmem176a
|
transmembrane protein 176A |
| chr11_+_120564185 | 1.32 |
ENSMUST00000135346.8
ENSMUST00000127269.8 ENSMUST00000131727.9 ENSMUST00000149389.8 ENSMUST00000153346.8 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr3_+_122039274 | 1.31 |
ENSMUST00000178826.8
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr17_-_24863907 | 1.29 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr1_+_165596961 | 1.24 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr18_+_61178211 | 1.21 |
ENSMUST00000025522.11
ENSMUST00000115274.2 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr10_-_95158827 | 1.21 |
ENSMUST00000220279.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr14_+_67953687 | 1.20 |
ENSMUST00000150768.8
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr11_+_69523754 | 1.20 |
ENSMUST00000018909.4
|
Fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr11_-_50101592 | 1.19 |
ENSMUST00000143379.2
ENSMUST00000015981.12 ENSMUST00000102774.11 |
Sqstm1
|
sequestosome 1 |
| chr10_+_101517348 | 1.19 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chrX_+_132809166 | 1.18 |
ENSMUST00000033606.15
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr11_+_50101717 | 1.16 |
ENSMUST00000147468.8
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
| chr4_+_117109204 | 1.14 |
ENSMUST00000125943.8
ENSMUST00000106434.8 |
Tmem53
|
transmembrane protein 53 |
| chr2_+_11710633 | 1.13 |
ENSMUST00000114832.3
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr1_-_155022501 | 1.13 |
ENSMUST00000027744.10
|
Mr1
|
major histocompatibility complex, class I-related |
| chr10_-_34294461 | 1.12 |
ENSMUST00000213269.2
ENSMUST00000099973.4 ENSMUST00000105512.8 ENSMUST00000047885.14 |
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr9_-_86577940 | 1.12 |
ENSMUST00000034989.15
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr8_-_68574179 | 1.11 |
ENSMUST00000178529.2
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr11_+_80319424 | 1.09 |
ENSMUST00000173938.8
ENSMUST00000017572.14 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr14_-_55950545 | 1.08 |
ENSMUST00000002389.9
ENSMUST00000226907.2 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr5_-_45607554 | 1.07 |
ENSMUST00000015950.12
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr19_-_8906686 | 1.06 |
ENSMUST00000096242.5
|
Rom1
|
rod outer segment membrane protein 1 |
| chrX_+_132809189 | 1.06 |
ENSMUST00000113304.2
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr8_+_106052970 | 1.06 |
ENSMUST00000015000.12
ENSMUST00000098453.9 |
Tmem208
|
transmembrane protein 208 |
| chr5_-_45607485 | 1.03 |
ENSMUST00000154962.8
ENSMUST00000118097.8 ENSMUST00000198258.5 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr15_-_101801351 | 1.03 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr9_+_5345450 | 1.02 |
ENSMUST00000151332.2
|
Casp12
|
caspase 12 |
| chr14_+_67953547 | 1.01 |
ENSMUST00000078053.13
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr5_-_73132045 | 1.00 |
ENSMUST00000043711.9
|
Gm10135
|
predicted gene 10135 |
| chr7_-_97794679 | 1.00 |
ENSMUST00000098281.4
|
Omp
|
olfactory marker protein |
| chr2_-_163261439 | 0.99 |
ENSMUST00000046908.10
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr7_-_113875261 | 0.94 |
ENSMUST00000135570.8
|
Psma1
|
proteasome subunit alpha 1 |
| chr19_+_8906916 | 0.94 |
ENSMUST00000096241.6
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr14_-_20443676 | 0.94 |
ENSMUST00000061444.5
|
Mrps16
|
mitochondrial ribosomal protein S16 |
| chr9_+_107456086 | 0.93 |
ENSMUST00000149638.8
ENSMUST00000139274.8 ENSMUST00000130053.8 ENSMUST00000122985.8 ENSMUST00000127380.8 ENSMUST00000139581.2 |
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr18_-_63825380 | 0.93 |
ENSMUST00000025476.4
|
Txnl1
|
thioredoxin-like 1 |
| chr3_+_108164242 | 0.92 |
ENSMUST00000090569.10
|
Psma5
|
proteasome subunit alpha 5 |
| chr12_-_83609217 | 0.91 |
ENSMUST00000222448.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr11_+_100510043 | 0.91 |
ENSMUST00000107376.8
|
Nkiras2
|
NFKB inhibitor interacting Ras-like protein 2 |
| chr9_-_116004265 | 0.87 |
ENSMUST00000061101.12
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr11_-_50216426 | 0.86 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr18_-_43610829 | 0.86 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr9_+_65121944 | 0.85 |
ENSMUST00000069000.9
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr9_+_5345405 | 0.84 |
ENSMUST00000027009.11
|
Casp12
|
caspase 12 |
| chr11_-_55075855 | 0.83 |
ENSMUST00000039305.6
|
Slc36a2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr11_-_78277384 | 0.82 |
ENSMUST00000108294.2
|
Foxn1
|
forkhead box N1 |
| chr16_+_20367327 | 0.82 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr2_+_151947444 | 0.81 |
ENSMUST00000041500.8
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr7_+_101875019 | 0.80 |
ENSMUST00000142873.8
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr5_-_113216632 | 0.80 |
ENSMUST00000031295.8
|
Crybb2
|
crystallin, beta B2 |
| chr9_-_21150100 | 0.78 |
ENSMUST00000164812.8
|
Keap1
|
kelch-like ECH-associated protein 1 |
| chr4_+_122910382 | 0.76 |
ENSMUST00000102649.4
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr10_+_127894816 | 0.75 |
ENSMUST00000052798.14
|
Ptges3
|
prostaglandin E synthase 3 |
| chr12_-_113760187 | 0.75 |
ENSMUST00000192911.2
ENSMUST00000103455.3 |
Ighv2-6-8
|
immunoglobulin heavy variable 2-6-8 |
| chr11_+_107370375 | 0.73 |
ENSMUST00000106750.5
|
Psmd12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr12_+_71021395 | 0.73 |
ENSMUST00000160027.8
ENSMUST00000160864.8 |
Psma3
|
proteasome subunit alpha 3 |
| chr2_+_153491363 | 0.72 |
ENSMUST00000072997.10
ENSMUST00000109773.8 ENSMUST00000109774.9 ENSMUST00000081628.13 ENSMUST00000103151.8 ENSMUST00000056495.14 ENSMUST00000088976.12 ENSMUST00000109772.8 ENSMUST00000103150.10 |
Dnmt3b
|
DNA methyltransferase 3B |
| chr10_+_127894843 | 0.70 |
ENSMUST00000084771.3
|
Ptges3
|
prostaglandin E synthase 3 |
| chr14_-_55950939 | 0.70 |
ENSMUST00000168729.8
ENSMUST00000228123.2 ENSMUST00000178034.9 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr9_-_116004386 | 0.70 |
ENSMUST00000035014.8
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr7_-_80475722 | 0.69 |
ENSMUST00000205304.2
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr11_+_76836545 | 0.68 |
ENSMUST00000125145.8
|
Blmh
|
bleomycin hydrolase |
| chr14_-_36857083 | 0.68 |
ENSMUST00000042564.17
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr11_+_107370303 | 0.68 |
ENSMUST00000021063.13
ENSMUST00000106752.10 |
Psmd12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr9_+_38120348 | 0.65 |
ENSMUST00000093867.3
|
Olfr893
|
olfactory receptor 893 |
| chr18_-_63825491 | 0.64 |
ENSMUST00000237004.2
|
Txnl1
|
thioredoxin-like 1 |
| chr11_+_74255497 | 0.64 |
ENSMUST00000077794.4
|
Olfr412
|
olfactory receptor 412 |
| chr7_-_113875342 | 0.64 |
ENSMUST00000033008.10
|
Psma1
|
proteasome subunit alpha 1 |
| chr7_+_141048722 | 0.64 |
ENSMUST00000058746.7
|
Cd151
|
CD151 antigen |
| chr10_+_101517556 | 0.62 |
ENSMUST00000156751.8
|
Mgat4c
|
MGAT4 family, member C |
| chr6_-_146403410 | 0.61 |
ENSMUST00000053273.15
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr14_-_77274056 | 0.61 |
ENSMUST00000062789.15
|
Lacc1
|
laccase domain containing 1 |
| chr19_+_29229147 | 0.61 |
ENSMUST00000025705.7
ENSMUST00000065796.10 ENSMUST00000236990.2 |
Jak2
|
Janus kinase 2 |
| chr5_-_45607463 | 0.61 |
ENSMUST00000197946.5
ENSMUST00000127562.3 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr1_+_36730530 | 0.61 |
ENSMUST00000081180.7
ENSMUST00000193210.6 ENSMUST00000195151.6 |
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr11_+_68779372 | 0.59 |
ENSMUST00000065213.5
|
Rnf222
|
ring finger protein 222 |
| chr11_+_4207557 | 0.58 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr7_-_103964662 | 0.57 |
ENSMUST00000106837.8
ENSMUST00000106839.9 ENSMUST00000070943.7 |
Trim12a
|
tripartite motif-containing 12A |
| chr14_+_54437843 | 0.57 |
ENSMUST00000103722.2
|
Traj19
|
T cell receptor alpha joining 19 |
| chr17_-_45884179 | 0.53 |
ENSMUST00000165127.8
ENSMUST00000166469.8 ENSMUST00000024739.14 |
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr9_+_32305259 | 0.51 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr2_+_163695571 | 0.51 |
ENSMUST00000044798.4
ENSMUST00000109396.2 |
Kcnk15
|
potassium channel, subfamily K, member 15 |
| chr1_-_130866978 | 0.50 |
ENSMUST00000016668.13
|
Il19
|
interleukin 19 |
| chr17_-_71305003 | 0.50 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr3_-_10248979 | 0.50 |
ENSMUST00000029034.9
|
Pmp2
|
peripheral myelin protein 2 |
| chr8_-_21392510 | 0.49 |
ENSMUST00000122025.9
|
Gm15056
|
predicted gene 15056 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 15.9 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 2.8 | 8.3 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 2.2 | 11.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 1.6 | 9.4 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.5 | 6.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 1.4 | 4.1 | GO:0014728 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 1.2 | 13.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.9 | 2.6 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.7 | 5.4 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.7 | 2.0 | GO:0015881 | creatine transport(GO:0015881) |
| 0.7 | 2.0 | GO:0002625 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.7 | 3.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.6 | 4.9 | GO:1901750 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.6 | 3.0 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.5 | 1.6 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of tolerance induction to self antigen(GO:0002649) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.5 | 4.0 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.5 | 2.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.4 | 1.7 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.4 | 1.2 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.4 | 1.9 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.4 | 4.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.4 | 8.9 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.3 | 1.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.3 | 2.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 1.7 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.3 | 0.8 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.3 | 2.7 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.3 | 3.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.3 | 1.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.3 | 4.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 12.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.3 | 0.8 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.2 | 0.7 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.2 | 1.4 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.2 | 7.6 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.2 | 1.4 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.2 | 1.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 1.5 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 0.8 | GO:0036233 | proline transmembrane transport(GO:0035524) glycine import(GO:0036233) |
| 0.2 | 0.8 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.2 | 3.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 0.6 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.2 | 4.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 1.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 1.7 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 | 1.7 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.2 | 2.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.2 | 2.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.5 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.2 | 2.2 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.2 | 1.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 1.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.6 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.1 | 0.4 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.1 | 0.9 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 6.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 4.0 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.1 | 3.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 6.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 1.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 2.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.3 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.6 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.7 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 1.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.5 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 1.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 4.6 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 1.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 3.8 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 5.8 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 1.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.4 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.5 | GO:0030950 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 4.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 2.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.3 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 2.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.0 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.0 | 6.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.0 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 2.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 4.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 1.4 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 4.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 1.1 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 1.4 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 1.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 3.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 3.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.3 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 3.3 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 3.0 | GO:0043434 | response to peptide hormone(GO:0043434) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 13.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 2.1 | 6.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.6 | 20.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.6 | 1.7 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.5 | 1.9 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.4 | 3.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 1.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.4 | 6.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 3.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 2.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 2.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 6.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 9.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 2.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 4.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 0.9 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.8 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 2.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 4.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.4 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 0.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 8.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.9 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 2.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.1 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 6.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 3.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 8.4 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 3.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 5.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 2.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0035731 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 3.4 | 13.6 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 1.0 | 6.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.9 | 9.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.9 | 2.7 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.7 | 2.0 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.7 | 3.3 | GO:0004854 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdopterin cofactor binding(GO:0043546) |
| 0.6 | 1.9 | GO:0008127 | quercetin 2,3-dioxygenase activity(GO:0008127) |
| 0.6 | 1.7 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.5 | 5.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.5 | 2.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.5 | 1.6 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.5 | 4.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 4.5 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.4 | 4.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 6.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.4 | 1.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.4 | 1.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 1.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 2.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 3.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 4.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 4.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.3 | 6.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.7 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.3 | 0.8 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.3 | 3.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 2.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 1.4 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 1.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 6.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.4 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.2 | 3.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.7 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.2 | 5.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 1.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 2.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 2.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.4 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.6 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.4 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.1 | 3.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 1.9 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.8 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 3.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 3.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 3.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 8.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 6.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 1.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 13.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 6.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 1.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 4.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 5.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 4.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 9.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 4.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.0 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 2.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 2.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 1.9 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 6.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 9.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 4.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 2.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 3.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 15.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 14.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 3.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 6.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 3.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 6.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 4.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 5.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 3.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 3.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 2.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 4.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |