Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nfia
Z-value: 2.82
Transcription factors associated with Nfia
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfia
|
ENSMUSG00000028565.19 | Nfia |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfia | mm39_v1_chr4_+_97665992_97666071 | 0.61 | 9.8e-09 | Click! |
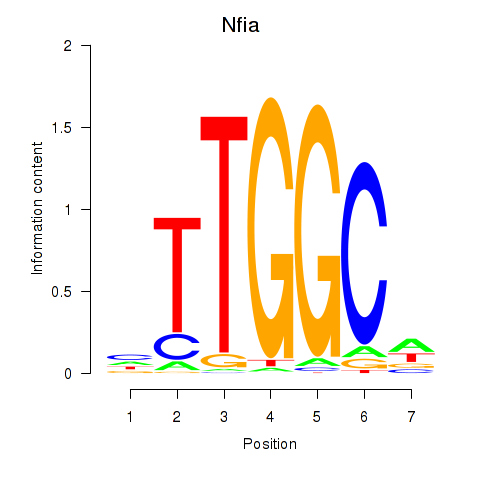
Activity profile of Nfia motif
Sorted Z-values of Nfia motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfia
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_140343652 | 18.50 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr18_+_20798337 | 16.59 |
ENSMUST00000075312.5
|
Ttr
|
transthyretin |
| chr7_-_140480314 | 13.75 |
ENSMUST00000026561.10
|
Cox8b
|
cytochrome c oxidase subunit 8B |
| chr6_-_115228800 | 13.17 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr7_-_97066937 | 12.80 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr2_-_113883285 | 12.78 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr7_+_130179063 | 12.14 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr11_-_5865124 | 12.03 |
ENSMUST00000109823.9
ENSMUST00000109822.8 |
Gck
|
glucokinase |
| chr15_-_78090591 | 12.03 |
ENSMUST00000120592.2
|
Pvalb
|
parvalbumin |
| chr7_-_48497771 | 11.89 |
ENSMUST00000032658.14
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr1_-_162726234 | 11.70 |
ENSMUST00000111510.8
ENSMUST00000045902.13 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr1_+_167445815 | 11.51 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr1_-_162687254 | 11.43 |
ENSMUST00000131058.8
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr6_+_90442269 | 11.06 |
ENSMUST00000113530.4
|
Klf15
|
Kruppel-like factor 15 |
| chr16_+_22710027 | 10.84 |
ENSMUST00000231848.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr11_-_75329726 | 10.73 |
ENSMUST00000108437.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr8_-_106670014 | 10.29 |
ENSMUST00000038896.8
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr4_+_123095297 | 10.26 |
ENSMUST00000068262.6
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr6_-_5193816 | 10.21 |
ENSMUST00000002663.12
|
Pon1
|
paraoxonase 1 |
| chr3_+_14928561 | 10.18 |
ENSMUST00000029076.6
|
Car3
|
carbonic anhydrase 3 |
| chr1_-_162687369 | 10.02 |
ENSMUST00000193078.6
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr17_+_32904629 | 9.93 |
ENSMUST00000008801.7
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr6_-_5193757 | 9.73 |
ENSMUST00000177159.9
ENSMUST00000176945.2 |
Pon1
|
paraoxonase 1 |
| chr17_+_32904601 | 9.49 |
ENSMUST00000168171.8
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr16_+_22710134 | 9.21 |
ENSMUST00000231328.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr6_-_54570124 | 9.20 |
ENSMUST00000046520.13
|
Fkbp14
|
FK506 binding protein 14 |
| chr11_+_78215026 | 9.16 |
ENSMUST00000102478.4
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr19_-_38113056 | 9.08 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr1_-_162726053 | 9.04 |
ENSMUST00000143123.3
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr11_+_78389913 | 9.03 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr4_+_137196080 | 8.79 |
ENSMUST00000030547.15
ENSMUST00000171332.2 |
Hspg2
|
perlecan (heparan sulfate proteoglycan 2) |
| chr15_-_3612703 | 8.72 |
ENSMUST00000069451.11
|
Ghr
|
growth hormone receptor |
| chr15_-_3612628 | 8.71 |
ENSMUST00000110698.9
|
Ghr
|
growth hormone receptor |
| chr4_-_118347249 | 8.66 |
ENSMUST00000047421.6
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr8_-_93857899 | 8.58 |
ENSMUST00000034189.17
|
Ces1c
|
carboxylesterase 1C |
| chr19_+_8828132 | 8.49 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr3_-_116762476 | 8.45 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr15_-_89024824 | 8.35 |
ENSMUST00000088827.8
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr3_+_137983250 | 8.26 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr8_+_96429665 | 8.23 |
ENSMUST00000073139.14
ENSMUST00000080666.8 ENSMUST00000212160.2 |
Ndrg4
|
N-myc downstream regulated gene 4 |
| chr13_-_19803928 | 8.15 |
ENSMUST00000221014.2
ENSMUST00000002885.8 ENSMUST00000220944.2 |
Epdr1
|
ependymin related protein 1 (zebrafish) |
| chr8_-_37200051 | 8.08 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr19_-_44396269 | 8.06 |
ENSMUST00000235741.2
|
Scd1
|
stearoyl-Coenzyme A desaturase 1 |
| chr19_-_44396092 | 7.96 |
ENSMUST00000041331.4
|
Scd1
|
stearoyl-Coenzyme A desaturase 1 |
| chr11_+_77353218 | 7.93 |
ENSMUST00000102493.8
|
Coro6
|
coronin 6 |
| chr15_+_54609011 | 7.92 |
ENSMUST00000050027.9
|
Ccn3
|
cellular communication network factor 3 |
| chr4_+_134042423 | 7.83 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr4_-_60777462 | 7.56 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr4_-_61700450 | 7.52 |
ENSMUST00000107477.2
ENSMUST00000080606.9 |
Mup19
|
major urinary protein 19 |
| chr2_-_164621641 | 7.43 |
ENSMUST00000103095.5
|
Tnnc2
|
troponin C2, fast |
| chr3_-_81883509 | 7.40 |
ENSMUST00000029645.14
ENSMUST00000193879.2 |
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr19_+_4644365 | 7.32 |
ENSMUST00000113825.4
|
Pcx
|
pyruvate carboxylase |
| chr5_-_31453206 | 7.24 |
ENSMUST00000041266.11
ENSMUST00000172435.8 ENSMUST00000201417.2 |
Fndc4
|
fibronectin type III domain containing 4 |
| chr4_-_61437704 | 7.23 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr4_+_99812912 | 7.18 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr4_-_60139857 | 7.18 |
ENSMUST00000107490.5
ENSMUST00000074700.9 |
Mup2
|
major urinary protein 2 |
| chr15_-_76906832 | 7.15 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr17_+_34783242 | 7.14 |
ENSMUST00000015612.14
|
Notch4
|
notch 4 |
| chr17_+_24946793 | 7.12 |
ENSMUST00000170239.9
|
Rpl3l
|
ribosomal protein L3-like |
| chr8_+_69333143 | 7.11 |
ENSMUST00000015712.15
|
Lpl
|
lipoprotein lipase |
| chr1_+_87522267 | 6.91 |
ENSMUST00000165109.2
ENSMUST00000070898.6 |
Neu2
|
neuraminidase 2 |
| chr2_-_163239865 | 6.88 |
ENSMUST00000017961.11
ENSMUST00000109425.3 |
Jph2
|
junctophilin 2 |
| chr15_-_96929086 | 6.84 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr3_+_137329433 | 6.81 |
ENSMUST00000053855.8
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr6_-_141801897 | 6.76 |
ENSMUST00000165990.8
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr15_-_41733099 | 6.74 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein |
| chr17_+_24947148 | 6.73 |
ENSMUST00000183214.2
|
Rpl3l
|
ribosomal protein L3-like |
| chr7_-_127805518 | 6.70 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr4_-_61259801 | 6.67 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr6_+_113460258 | 6.66 |
ENSMUST00000032422.6
|
Creld1
|
cysteine-rich with EGF-like domains 1 |
| chr6_-_53797748 | 6.65 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr18_+_44467133 | 6.59 |
ENSMUST00000025349.12
ENSMUST00000115498.2 |
Myot
|
myotilin |
| chr7_-_105249308 | 6.57 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr7_-_25358406 | 6.55 |
ENSMUST00000071329.8
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr18_+_32055339 | 6.54 |
ENSMUST00000233994.2
|
Lims2
|
LIM and senescent cell antigen like domains 2 |
| chr11_-_61269131 | 6.53 |
ENSMUST00000148671.2
|
Slc47a1
|
solute carrier family 47, member 1 |
| chr12_+_104304631 | 6.52 |
ENSMUST00000043058.5
ENSMUST00000101078.12 |
Serpina3k
Serpina3m
|
serine (or cysteine) peptidase inhibitor, clade A, member 3K serine (or cysteine) peptidase inhibitor, clade A, member 3M |
| chr13_+_49658249 | 6.50 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr7_+_51528788 | 6.47 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr9_+_110782861 | 6.46 |
ENSMUST00000196834.2
|
Lrrc2
|
leucine rich repeat containing 2 |
| chr14_-_61283911 | 6.45 |
ENSMUST00000111234.10
ENSMUST00000224371.2 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr16_+_6166982 | 6.41 |
ENSMUST00000056416.9
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr6_-_129428746 | 6.38 |
ENSMUST00000204012.2
ENSMUST00000037481.10 |
Clec1a
|
C-type lectin domain family 1, member a |
| chr6_-_141801918 | 6.34 |
ENSMUST00000163678.2
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr2_+_151985467 | 6.33 |
ENSMUST00000089112.6
|
Tcf15
|
transcription factor 15 |
| chr5_+_67125759 | 6.31 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr1_-_66984178 | 6.26 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr1_+_152275575 | 6.24 |
ENSMUST00000044311.9
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr1_-_172047282 | 6.16 |
ENSMUST00000170700.2
ENSMUST00000003554.11 |
Casq1
|
calsequestrin 1 |
| chrX_-_20816841 | 6.13 |
ENSMUST00000009550.14
|
Elk1
|
ELK1, member of ETS oncogene family |
| chr7_+_119217004 | 6.11 |
ENSMUST00000047929.13
ENSMUST00000135683.3 |
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr4_+_105014536 | 6.10 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr3_+_145464413 | 6.02 |
ENSMUST00000029845.15
|
Ddah1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr14_+_66208059 | 6.01 |
ENSMUST00000127387.8
|
Clu
|
clusterin |
| chr1_+_140173787 | 6.00 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr9_+_21095399 | 5.98 |
ENSMUST00000115458.9
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr5_-_35828857 | 5.83 |
ENSMUST00000129459.2
ENSMUST00000137935.2 |
Htra3
|
HtrA serine peptidase 3 |
| chr9_-_48747232 | 5.82 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr16_+_22713593 | 5.81 |
ENSMUST00000232674.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr7_+_126396779 | 5.81 |
ENSMUST00000205324.2
|
Tlcd3b
|
TLC domain containing 3B |
| chr4_+_95467701 | 5.80 |
ENSMUST00000150830.2
ENSMUST00000134012.9 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr8_-_46664321 | 5.80 |
ENSMUST00000034049.5
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr2_-_71198091 | 5.79 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr19_+_16933471 | 5.74 |
ENSMUST00000087689.5
|
Prune2
|
prune homolog 2 |
| chr14_-_70761507 | 5.73 |
ENSMUST00000022692.5
|
Sftpc
|
surfactant associated protein C |
| chr9_+_98305014 | 5.71 |
ENSMUST00000052068.11
|
Rbp1
|
retinol binding protein 1, cellular |
| chr18_-_62044871 | 5.69 |
ENSMUST00000166783.3
ENSMUST00000049378.15 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr14_+_66207163 | 5.69 |
ENSMUST00000153460.8
|
Clu
|
clusterin |
| chr6_+_90310252 | 5.68 |
ENSMUST00000046128.12
ENSMUST00000164761.6 |
Uroc1
|
urocanase domain containing 1 |
| chr11_+_52265090 | 5.68 |
ENSMUST00000020673.3
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr19_+_4644425 | 5.66 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr6_-_115229128 | 5.65 |
ENSMUST00000032462.9
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr11_+_67128843 | 5.65 |
ENSMUST00000018632.11
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr8_-_71848429 | 5.64 |
ENSMUST00000049184.9
|
Ushbp1
|
USH1 protein network component harmonin binding protein 1 |
| chr2_+_164404499 | 5.60 |
ENSMUST00000017867.10
ENSMUST00000109344.9 ENSMUST00000109345.9 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr6_+_21986445 | 5.59 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr6_-_129428869 | 5.58 |
ENSMUST00000203162.3
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr3_+_82915031 | 5.49 |
ENSMUST00000048486.13
ENSMUST00000194175.2 |
Fgg
|
fibrinogen gamma chain |
| chr7_+_89281897 | 5.48 |
ENSMUST00000032856.13
|
Me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr9_+_108356935 | 5.47 |
ENSMUST00000194147.2
ENSMUST00000065014.10 ENSMUST00000195483.6 ENSMUST00000195058.2 |
Lamb2
|
laminin, beta 2 |
| chr16_+_20513658 | 5.47 |
ENSMUST00000056518.13
|
Fam131a
|
family with sequence similarity 131, member A |
| chr6_-_124410452 | 5.47 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr5_-_52628825 | 5.42 |
ENSMUST00000198008.5
ENSMUST00000059428.7 |
Ccdc149
|
coiled-coil domain containing 149 |
| chr11_+_98274637 | 5.41 |
ENSMUST00000008021.3
|
Tcap
|
titin-cap |
| chr14_-_121935829 | 5.40 |
ENSMUST00000040700.9
ENSMUST00000212181.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr4_-_61972348 | 5.38 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr2_+_172994841 | 5.33 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr3_+_99203818 | 5.32 |
ENSMUST00000150756.3
|
Tbx15
|
T-box 15 |
| chr11_+_28803188 | 5.30 |
ENSMUST00000020759.12
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chr5_-_147662798 | 5.29 |
ENSMUST00000110529.6
ENSMUST00000031653.12 |
Flt1
|
FMS-like tyrosine kinase 1 |
| chr2_+_119181703 | 5.27 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr11_+_98938137 | 5.25 |
ENSMUST00000140772.2
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr9_-_48747474 | 5.24 |
ENSMUST00000216150.2
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr18_-_35788255 | 5.24 |
ENSMUST00000190196.5
|
Prob1
|
proline rich basic protein 1 |
| chr7_-_45138188 | 5.22 |
ENSMUST00000011526.7
|
Dhdh
|
dihydrodiol dehydrogenase (dimeric) |
| chr14_+_19801333 | 5.21 |
ENSMUST00000022340.5
|
Nid2
|
nidogen 2 |
| chr1_-_135302971 | 5.21 |
ENSMUST00000041240.4
|
Shisa4
|
shisa family member 4 |
| chr17_+_36132567 | 5.17 |
ENSMUST00000003635.7
|
Ier3
|
immediate early response 3 |
| chr15_+_32920869 | 5.16 |
ENSMUST00000022871.7
|
Sdc2
|
syndecan 2 |
| chr17_-_37334240 | 5.16 |
ENSMUST00000102665.11
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr9_-_26717686 | 5.16 |
ENSMUST00000162702.8
ENSMUST00000040398.14 ENSMUST00000066560.13 |
Glb1l2
|
galactosidase, beta 1-like 2 |
| chrX_+_156485570 | 5.13 |
ENSMUST00000112520.2
|
Smpx
|
small muscle protein, X-linked |
| chr19_+_5790918 | 5.12 |
ENSMUST00000081496.6
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr8_+_47439948 | 5.02 |
ENSMUST00000119686.2
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr13_+_49761506 | 5.02 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr1_-_134162231 | 4.99 |
ENSMUST00000169927.2
|
Adora1
|
adenosine A1 receptor |
| chr9_-_103099262 | 4.99 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr12_-_91712783 | 4.96 |
ENSMUST00000166967.2
|
Ston2
|
stonin 2 |
| chr16_+_41353212 | 4.96 |
ENSMUST00000078873.11
|
Lsamp
|
limbic system-associated membrane protein |
| chr7_+_140425460 | 4.91 |
ENSMUST00000035300.7
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr5_-_115332343 | 4.91 |
ENSMUST00000112113.8
|
Cabp1
|
calcium binding protein 1 |
| chr17_-_26727437 | 4.91 |
ENSMUST00000236661.2
ENSMUST00000025025.7 |
Dusp1
|
dual specificity phosphatase 1 |
| chr2_+_32536594 | 4.90 |
ENSMUST00000113272.8
ENSMUST00000009705.14 ENSMUST00000167841.8 |
Eng
|
endoglin |
| chr11_+_75400889 | 4.89 |
ENSMUST00000042972.7
|
Rilp
|
Rab interacting lysosomal protein |
| chr3_+_137329791 | 4.89 |
ENSMUST00000165845.2
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr7_-_44174065 | 4.89 |
ENSMUST00000165208.4
|
Mybpc2
|
myosin binding protein C, fast-type |
| chrX_+_163219983 | 4.89 |
ENSMUST00000036858.11
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr2_-_154249997 | 4.87 |
ENSMUST00000028991.7
ENSMUST00000109728.8 |
Snta1
|
syntrophin, acidic 1 |
| chr14_-_66361931 | 4.86 |
ENSMUST00000070515.2
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr1_-_163552693 | 4.84 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr1_+_134121170 | 4.84 |
ENSMUST00000038445.13
ENSMUST00000191577.2 |
Mybph
|
myosin binding protein H |
| chr17_-_48739874 | 4.82 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr15_-_11905697 | 4.82 |
ENSMUST00000066529.5
ENSMUST00000228603.2 |
Npr3
|
natriuretic peptide receptor 3 |
| chr11_+_49500090 | 4.80 |
ENSMUST00000020617.3
|
Flt4
|
FMS-like tyrosine kinase 4 |
| chr5_-_87288177 | 4.80 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr13_-_32960379 | 4.79 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr1_-_189901596 | 4.78 |
ENSMUST00000010319.14
|
Prox1
|
prospero homeobox 1 |
| chr6_+_141470105 | 4.77 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr5_+_24569802 | 4.72 |
ENSMUST00000115090.6
ENSMUST00000030834.7 |
Nos3
|
nitric oxide synthase 3, endothelial cell |
| chr11_-_121279062 | 4.71 |
ENSMUST00000106107.3
|
Rab40b
|
Rab40B, member RAS oncogene family |
| chr15_-_39720855 | 4.70 |
ENSMUST00000022915.11
ENSMUST00000110306.9 |
Dpys
|
dihydropyrimidinase |
| chr5_+_16139760 | 4.70 |
ENSMUST00000101581.10
ENSMUST00000039370.14 ENSMUST00000199704.5 ENSMUST00000180204.8 ENSMUST00000078272.13 ENSMUST00000115281.7 |
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr1_+_93163553 | 4.68 |
ENSMUST00000062202.14
|
Sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr1_-_74788013 | 4.65 |
ENSMUST00000188073.7
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr9_+_54824547 | 4.59 |
ENSMUST00000039742.8
|
Hykk
|
hydroxylysine kinase 1 |
| chr3_+_137046828 | 4.59 |
ENSMUST00000122064.8
ENSMUST00000119475.6 |
Emcn
|
endomucin |
| chrX_+_37405054 | 4.58 |
ENSMUST00000016471.9
ENSMUST00000115134.2 |
Atp1b4
|
ATPase, (Na+)/K+ transporting, beta 4 polypeptide |
| chr11_-_83959380 | 4.58 |
ENSMUST00000164891.8
|
Dusp14
|
dual specificity phosphatase 14 |
| chr16_-_85599994 | 4.57 |
ENSMUST00000023610.15
|
Adamts1
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 |
| chrX_-_151820545 | 4.57 |
ENSMUST00000051484.5
|
Mageh1
|
MAGE family member H1 |
| chr15_-_37792635 | 4.57 |
ENSMUST00000090150.11
ENSMUST00000150453.2 |
Ncald
|
neurocalcin delta |
| chr5_+_90608751 | 4.56 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr7_+_18810097 | 4.54 |
ENSMUST00000032570.14
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr17_+_64170967 | 4.53 |
ENSMUST00000232945.2
|
Fer
|
fer (fms/fps related) protein kinase |
| chr13_+_16189041 | 4.49 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chr6_-_136852792 | 4.48 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chrX_-_51702790 | 4.47 |
ENSMUST00000069360.14
|
Gpc3
|
glypican 3 |
| chr18_-_35788240 | 4.45 |
ENSMUST00000097619.2
|
Prob1
|
proline rich basic protein 1 |
| chrX_+_165127688 | 4.45 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr4_+_148085179 | 4.44 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr18_-_33597060 | 4.44 |
ENSMUST00000168890.2
|
Nrep
|
neuronal regeneration related protein |
| chr18_-_33596890 | 4.42 |
ENSMUST00000237066.2
|
Nrep
|
neuronal regeneration related protein |
| chr14_+_31750946 | 4.41 |
ENSMUST00000022460.11
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chr19_+_4608836 | 4.41 |
ENSMUST00000236457.2
|
Pcx
|
pyruvate carboxylase |
| chr1_+_58152295 | 4.39 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr15_-_37458768 | 4.36 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr9_+_106339069 | 4.35 |
ENSMUST00000188396.2
|
Pcbp4
|
poly(rC) binding protein 4 |
| chr3_+_130411097 | 4.34 |
ENSMUST00000166187.8
ENSMUST00000072271.13 |
Etnppl
|
ethanolamine phosphate phospholyase |
| chr8_+_71207326 | 4.29 |
ENSMUST00000110093.9
ENSMUST00000143118.3 ENSMUST00000034301.12 ENSMUST00000110090.8 |
Rab3a
|
RAB3A, member RAS oncogene family |
| chr12_+_75355082 | 4.28 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.9 | GO:1902617 | response to fluoride(GO:1902617) |
| 5.7 | 5.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 5.3 | 16.0 | GO:1903699 | tarsal gland development(GO:1903699) |
| 5.1 | 5.1 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 4.9 | 4.9 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 4.0 | 11.9 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 3.5 | 17.4 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 3.5 | 20.7 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 2.8 | 2.8 | GO:0048389 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 2.7 | 8.0 | GO:0072312 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 2.6 | 7.8 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 2.5 | 7.4 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 2.4 | 12.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 2.4 | 19.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 2.2 | 8.9 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 2.1 | 23.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 2.1 | 8.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 2.1 | 14.4 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 1.9 | 11.7 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.9 | 7.6 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 1.8 | 9.2 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.8 | 9.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 1.8 | 8.9 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.8 | 7.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 1.7 | 5.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 1.7 | 8.6 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 1.7 | 5.0 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 1.7 | 5.0 | GO:0042323 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.6 | 4.9 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 1.6 | 6.5 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 1.6 | 19.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.6 | 9.5 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.6 | 4.7 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 1.6 | 4.7 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 1.6 | 25.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.5 | 13.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 1.5 | 6.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.5 | 4.6 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 1.5 | 4.4 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 1.5 | 5.9 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 1.5 | 5.8 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.5 | 10.2 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 1.4 | 39.1 | GO:0017144 | drug metabolic process(GO:0017144) |
| 1.4 | 4.3 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 1.4 | 12.7 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.4 | 4.1 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.3 | 4.0 | GO:2000722 | response to mineralocorticoid(GO:0051385) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 1.3 | 14.8 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 1.3 | 10.7 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 1.3 | 17.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 1.3 | 4.0 | GO:0060365 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 1.3 | 6.6 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 1.2 | 2.5 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 1.2 | 11.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.2 | 2.4 | GO:0021586 | pons maturation(GO:0021586) |
| 1.2 | 10.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.2 | 3.5 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.2 | 5.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.1 | 3.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 1.1 | 5.7 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 1.1 | 3.4 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 1.1 | 5.6 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.1 | 4.4 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 1.1 | 5.4 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 1.1 | 3.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 1.1 | 4.2 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 1.0 | 2.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 1.0 | 7.1 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 1.0 | 3.1 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 1.0 | 4.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 1.0 | 3.0 | GO:0038190 | cell migration involved in vasculogenesis(GO:0035441) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 1.0 | 3.9 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.0 | 2.9 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 1.0 | 2.9 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.0 | 1.9 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.0 | 3.8 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.9 | 4.7 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.9 | 6.6 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.9 | 3.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.9 | 7.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.9 | 3.7 | GO:0006507 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.9 | 2.8 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.9 | 4.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.9 | 5.4 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.9 | 9.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.9 | 4.4 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.9 | 5.2 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.9 | 6.0 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.9 | 6.9 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.9 | 5.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.8 | 2.5 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) |
| 0.8 | 4.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.8 | 3.3 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.8 | 2.5 | GO:0030070 | insulin processing(GO:0030070) |
| 0.8 | 4.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.8 | 6.5 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.8 | 4.9 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.8 | 2.4 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.8 | 2.4 | GO:2000729 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.8 | 3.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.8 | 2.4 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.8 | 2.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.8 | 0.8 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.8 | 5.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.8 | 3.8 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.8 | 17.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.7 | 4.5 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.7 | 2.9 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.7 | 5.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.7 | 25.3 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.7 | 3.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.7 | 8.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.7 | 9.0 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.7 | 10.3 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.7 | 2.7 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.7 | 4.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.7 | 6.6 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.7 | 5.3 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.7 | 5.2 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.7 | 2.6 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.7 | 4.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.7 | 3.9 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.6 | 5.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.6 | 9.0 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.6 | 21.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.6 | 1.9 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.6 | 2.5 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.6 | 3.1 | GO:0046618 | drug export(GO:0046618) |
| 0.6 | 4.9 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.6 | 2.4 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.6 | 6.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.6 | 5.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.6 | 1.2 | GO:1904708 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 0.6 | 1.8 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.6 | 6.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.6 | 1.8 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.6 | 2.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.6 | 6.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.6 | 7.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.6 | 1.7 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.6 | 3.5 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.6 | 4.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.6 | 6.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.6 | 1.7 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.6 | 4.0 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.6 | 8.4 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.6 | 5.6 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.6 | 3.9 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.6 | 2.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.6 | 5.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.5 | 13.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.5 | 12.6 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.5 | 4.8 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.5 | 5.3 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.5 | 9.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.5 | 2.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.5 | 3.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.5 | 12.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.5 | 6.1 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 4.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.5 | 2.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.5 | 16.8 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.5 | 4.8 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.5 | 2.4 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.5 | 1.9 | GO:0015755 | fructose transport(GO:0015755) |
| 0.5 | 2.8 | GO:0097117 | protein complex assembly involved in synapse maturation(GO:0090126) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) guanylate kinase-associated protein clustering(GO:0097117) |
| 0.5 | 6.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.5 | 1.4 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.5 | 5.5 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.5 | 12.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.5 | 4.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 | 3.6 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.4 | 1.8 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.4 | 1.3 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.4 | 0.4 | GO:0060067 | cervix development(GO:0060067) |
| 0.4 | 1.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 0.9 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.4 | 1.3 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.4 | 3.9 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.4 | 2.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 2.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.4 | 5.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 8.0 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.4 | 18.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.4 | 5.4 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.4 | 11.1 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.4 | 4.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.4 | 2.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.4 | 5.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 1.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.4 | 1.2 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.4 | 1.2 | GO:0072049 | comma-shaped body morphogenesis(GO:0072049) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.4 | 2.4 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.4 | 1.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.4 | 1.5 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.4 | 5.4 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.4 | 2.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.4 | 1.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.4 | 4.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 2.6 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.4 | 0.7 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.4 | 1.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.4 | 4.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 7.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.4 | 7.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.4 | 2.9 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.4 | 3.6 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 4.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.4 | 3.2 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.4 | 8.8 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.4 | 1.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.3 | 1.0 | GO:0015881 | creatine transport(GO:0015881) |
| 0.3 | 11.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.3 | 5.2 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.3 | 1.7 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.3 | 2.4 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.3 | 5.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.3 | 2.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 | 8.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 2.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.3 | 3.7 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.3 | 7.3 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) |
| 0.3 | 1.6 | GO:1904720 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 | 0.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.3 | 4.0 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 2.7 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.3 | 3.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.3 | 5.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.3 | 1.8 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.3 | 2.9 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 0.9 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.3 | 4.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.3 | 0.9 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.3 | 1.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.3 | 2.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.4 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.3 | 3.5 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.3 | 4.4 | GO:0036296 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) |
| 0.3 | 3.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 4.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.3 | 2.7 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.3 | 10.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 1.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.3 | 2.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 3.9 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.3 | 6.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.3 | 1.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.3 | 1.8 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.3 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 1.5 | GO:0097688 | glutamate receptor clustering(GO:0097688) |
| 0.3 | 6.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 10.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 2.2 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.2 | 1.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 9.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.7 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.2 | 3.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 2.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.2 | 1.2 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.2 | 0.9 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.2 | 1.6 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 | 1.8 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.2 | 0.5 | GO:0032417 | regulation of sodium:proton antiporter activity(GO:0032415) positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.2 | 1.4 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.2 | 4.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.2 | 8.8 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.2 | 1.6 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 0.4 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.2 | 1.3 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.2 | 0.7 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 1.1 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) negative regulation of defense response to bacterium(GO:1900425) |
| 0.2 | 1.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 0.6 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.2 | 10.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 9.2 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.2 | 2.3 | GO:0097286 | iron ion import(GO:0097286) |
| 0.2 | 2.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 0.8 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.2 | 9.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 2.3 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.2 | 3.9 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 1.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 0.9 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.2 | 7.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 1.1 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.2 | 0.7 | GO:0070781 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.2 | 20.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.2 | 2.2 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.2 | 1.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 6.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 2.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 3.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 0.5 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 0.7 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.2 | 0.5 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 2.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 5.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 9.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.2 | 3.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.2 | 3.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 3.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 2.9 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.2 | 1.0 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 6.3 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.2 | 7.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 0.8 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 0.6 | GO:0000966 | pachytene(GO:0000239) RNA 5'-end processing(GO:0000966) |
| 0.2 | 6.8 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.2 | 0.6 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.2 | 0.9 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 1.1 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.2 | 1.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 5.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 7.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 4.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 3.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 2.6 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 9.0 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 0.6 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 1.6 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 7.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 3.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.7 | GO:0090179 | regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 5.9 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.1 | 1.4 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.1 | 1.3 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.6 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 2.1 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.1 | 4.9 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 11.7 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 1.7 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 1.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.4 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.1 | 1.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 17.0 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 0.9 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 2.3 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 3.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 2.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.9 | GO:0015893 | drug transport(GO:0015893) |
| 0.1 | 2.9 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 1.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.0 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 6.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.1 | 0.6 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 2.1 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 2.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.8 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 1.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 1.7 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 2.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.1 | 1.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 2.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.8 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 3.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 1.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.7 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.3 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.2 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.1 | 0.7 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.6 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 10.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 2.0 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 1.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 1.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 1.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.9 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 1.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 1.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 2.9 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 7.5 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.1 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.5 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 1.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.5 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.1 | 1.6 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 8.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 1.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.4 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.9 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.5 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.1 | 0.7 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 6.1 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.1 | 0.8 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.1 | 0.5 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 3.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.2 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.1 | 0.9 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 1.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 8.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 0.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 10.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.8 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 1.8 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 3.4 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 1.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 1.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 1.7 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.1 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.5 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 2.6 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 1.3 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.5 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 1.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 2.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.7 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.1 | 2.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.6 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of developmental pigmentation(GO:0048087) positive regulation of pigment cell differentiation(GO:0050942) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.1 | 1.8 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.7 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.1 | 2.7 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 2.1 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 3.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 2.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 4.0 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 1.5 | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation(GO:0060393) |
| 0.1 | 0.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.4 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 1.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.6 | GO:0060479 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.9 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 3.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 2.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.8 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.9 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.1 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.7 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 4.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.2 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 7.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 45.7 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.6 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.7 | GO:0000423 | macromitophagy(GO:0000423) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 1.0 | GO:0003143 | heart looping(GO:0001947) embryonic heart tube morphogenesis(GO:0003143) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.1 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 1.4 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 1.0 | GO:1904377 | positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 1.6 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 1.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 1.8 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.6 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 3.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.5 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.9 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 1.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.5 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 1.4 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.4 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.1 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 4.3 | 12.8 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 3.1 | 9.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 2.0 | 29.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.9 | 7.7 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 1.8 | 16.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.6 | 9.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 1.6 | 10.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.4 | 5.5 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.3 | 8.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.3 | 25.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.3 | 3.9 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.3 | 3.9 | GO:0043512 | inhibin A complex(GO:0043512) |
| 1.2 | 6.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.0 | 4.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.0 | 11.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 1.0 | 7.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.9 | 8.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.9 | 4.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.8 | 6.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.8 | 4.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.8 | 7.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.8 | 19.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.8 | 5.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.8 | 3.8 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.7 | 2.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.6 | 5.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.6 | 3.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.6 | 10.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.6 | 7.4 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.6 | 2.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.6 | 1.8 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.6 | 2.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.6 | 11.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.6 | 2.9 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.6 | 2.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.6 | 13.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.5 | 5.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.5 | 13.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 5.6 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 6.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.5 | 2.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.5 | 5.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.5 | 4.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 1.8 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.4 | 6.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.4 | 5.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 4.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.4 | 3.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.4 | 1.7 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.4 | 9.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.4 | 7.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 4.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 1.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 1.2 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.4 | 5.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 5.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 1.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.4 | 2.9 | GO:0001652 | granular component(GO:0001652) |
| 0.4 | 1.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.4 | 2.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 13.6 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 8.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 1.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.3 | 8.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 1.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.3 | 4.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 2.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 1.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 1.1 | GO:0044301 | climbing fiber(GO:0044301) parallel fiber(GO:1990032) |
| 0.3 | 1.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 5.3 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.3 | 4.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 4.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 2.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 2.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 3.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 3.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 0.7 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 1.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 85.8 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.2 | 3.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 3.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 3.6 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 5.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 4.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 54.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 1.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 2.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 12.5 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.2 | 1.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 20.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.2 | 3.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 4.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 2.7 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 0.6 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.2 | 1.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 2.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 4.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 4.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 4.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 1.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 6.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 34.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.2 | 1.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 2.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 6.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 3.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 21.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 6.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 4.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 1.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 4.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 4.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 2.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 2.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 2.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 4.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 7.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.5 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 0.8 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 3.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 18.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 8.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 53.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 3.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.5 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 10.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 8.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 9.4 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 7.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 5.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 21.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 5.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 2.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 19.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 5.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.6 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 4.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 9.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.7 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 91.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.1 | 2.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.5 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 7.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.2 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 4.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 50.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 5.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.7 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.5 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.5 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 3.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 6.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 5.9 | GO:0098791 | organelle subcompartment(GO:0031984) Golgi subcompartment(GO:0098791) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 23.7 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 5.8 | 17.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 4.3 | 17.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 4.3 | 25.9 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 3.7 | 44.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 3.6 | 21.8 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 3.2 | 9.7 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 3.0 | 9.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 2.9 | 8.6 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 2.8 | 8.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 2.6 | 7.8 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 2.2 | 15.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 2.2 | 6.5 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 1.9 | 11.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.9 | 3.8 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 1.9 | 13.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 1.9 | 5.6 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.9 | 7.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.8 | 5.5 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.8 | 5.3 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.7 | 10.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.6 | 4.9 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.6 | 6.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.6 | 9.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.6 | 4.7 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 1.6 | 4.7 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.5 | 4.6 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.5 | 10.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 1.4 | 5.6 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 1.4 | 6.9 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 1.4 | 4.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 1.4 | 19.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.3 | 4.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 1.3 | 5.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.3 | 6.5 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 1.3 | 5.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 1.2 | 7.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 1.2 | 8.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.2 | 12.0 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 1.2 | 7.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.2 | 4.7 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 1.2 | 10.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.2 | 5.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.2 | 11.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.1 | 3.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.1 | 3.4 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 1.1 | 11.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.1 | 4.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 1.1 | 4.5 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 1.1 | 5.6 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 1.1 | 25.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 1.1 | 4.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 1.1 | 6.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.1 | 5.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.1 | 4.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 1.0 | 7.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.0 | 4.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.0 | 22.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.0 | 3.0 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 1.0 | 3.0 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 1.0 | 4.9 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.9 | 26.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.9 | 1.8 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.9 | 3.6 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.9 | 8.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.9 | 4.3 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.8 | 7.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.8 | 8.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.8 | 4.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.8 | 3.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.8 | 10.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.8 | 2.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.8 | 6.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.7 | 2.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.7 | 2.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.7 | 2.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.7 | 5.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.7 | 7.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.7 | 6.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 4.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.7 | 11.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.6 | 3.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.6 | 2.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.6 | 1.9 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) uniporter activity(GO:0015292) |
| 0.6 | 20.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.6 | 2.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.6 | 4.9 | GO:0005534 | galactose binding(GO:0005534) |
| 0.6 | 14.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.6 | 9.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.6 | 2.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.6 | 3.5 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.6 | 2.9 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.6 | 1.7 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.6 | 11.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.6 | 5.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.6 | 8.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.5 | 6.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.5 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.5 | 5.4 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.5 | 4.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.5 | 10.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.5 | 2.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.5 | 6.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 6.5 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.5 | 4.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.5 | 4.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.5 | 3.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.5 | 9.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 4.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.5 | 3.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.4 | 14.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.4 | 5.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 2.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 4.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.4 | 10.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 7.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 3.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.4 | 7.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 8.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.4 | 4.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.4 | 1.6 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.4 | 3.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 7.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 2.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.4 | 1.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.4 | 5.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.4 | 1.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.4 | 1.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.4 | 4.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 4.6 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.4 | 1.1 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.4 | 2.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 3.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 4.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 1.0 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.3 | 3.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.3 | 2.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 6.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 1.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.3 | 8.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 11.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 1.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 2.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 3.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 1.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 13.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 4.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 0.9 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.3 | 4.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 12.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 2.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 1.8 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 2.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 12.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 7.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.3 | 3.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.3 | 1.7 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.3 | 5.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.3 | 0.8 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.3 | 1.1 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.3 | 0.8 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.3 | 1.4 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.3 | 7.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 7.0 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 5.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 6.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 1.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 4.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.3 | 4.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 12.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 1.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.3 | 3.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 1.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 2.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 2.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 8.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 2.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 2.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 0.7 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 4.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 3.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 3.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 4.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 2.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.2 | 5.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 4.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 3.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 2.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 4.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 2.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 1.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 7.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 22.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 14.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.6 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.2 | 2.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 1.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 8.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 2.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 3.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 2.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 1.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 1.3 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.2 | 0.7 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 0.7 | GO:0004079 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.2 | 8.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 8.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 0.5 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.2 | 6.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 1.9 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 4.6 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.2 | 16.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 1.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 0.5 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 1.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 1.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 0.3 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.2 | 12.2 | GO:0020037 | heme binding(GO:0020037) |
| 0.2 | 8.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 0.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 24.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 5.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 0.8 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 4.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 8.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.0 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 8.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 2.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 4.4 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.1 | 1.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 5.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.0 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 13.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 2.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 3.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 2.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 11.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 3.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 3.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 9.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 2.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 77.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.9 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.3 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.1 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.4 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 1.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 3.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 4.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 3.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 4.5 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 10.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 8.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 9.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 1.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 8.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 3.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.0 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.7 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 8.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.5 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 31.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 1.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.9 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 0.4 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 2.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 18.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 2.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 7.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 4.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 4.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 5.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 8.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 3.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 27.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.9 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 1.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 1.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 4.0 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 2.5 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 18.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.7 | 11.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.6 | 51.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 33.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.5 | 2.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 20.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 16.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.4 | 18.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 10.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.3 | 5.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 63.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 19.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.3 | 4.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 64.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 9.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 2.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 9.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 4.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 37.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 4.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 4.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 12.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 5.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 7.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 6.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 7.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 8.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 5.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 8.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 6.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 2.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 6.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 5.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 3.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 7.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 5.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 3.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 2.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 8.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 1.7 | 16.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 1.0 | 13.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.9 | 22.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.9 | 17.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.7 | 21.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.7 | 7.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.7 | 9.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.7 | 42.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.7 | 20.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.6 | 7.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.6 | 13.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.6 | 20.3 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.5 | 15.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.5 | 7.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.5 | 10.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 5.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 7.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.5 | 16.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 8.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.4 | 8.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.4 | 25.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 19.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 3.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 12.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.4 | 11.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 4.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.4 | 3.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 5.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 6.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 4.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 3.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 8.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.3 | 2.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.3 | 4.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 5.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 2.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 2.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.3 | 10.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 10.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 3.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 6.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 12.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 3.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 6.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 7.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 9.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 14.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.3 | 2.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 8.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 5.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 4.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 10.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 14.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 3.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 2.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 1.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 2.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 7.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 12.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 4.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 6.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 3.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 4.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 1.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 13.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 2.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 4.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.3 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 2.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 5.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 1.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 5.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 4.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.9 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 2.2 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 3.6 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 15.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 4.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 3.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 2.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 3.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 3.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.0 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |