Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
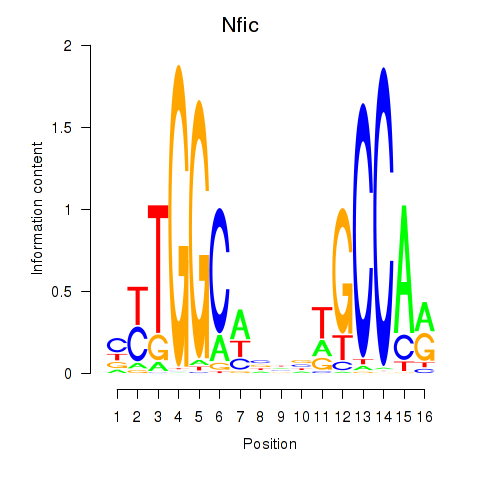
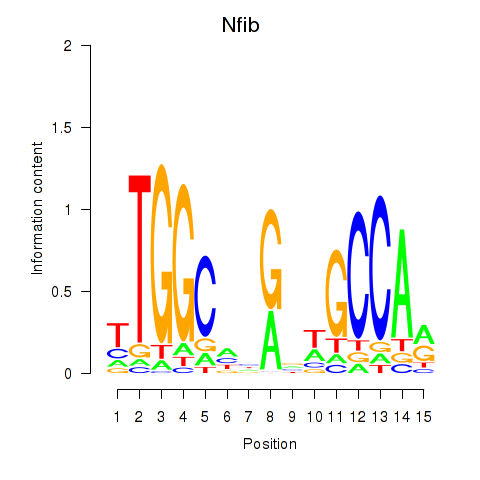
Results for Nfic_Nfib
Z-value: 2.07
Transcription factors associated with Nfic_Nfib
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfic
|
ENSMUSG00000055053.19 | Nfic |
|
Nfib
|
ENSMUSG00000008575.18 | Nfib |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfib | mm39_v1_chr4_-_82423944_82423978 | 0.75 | 3.8e-14 | Click! |
| Nfic | mm39_v1_chr10_-_81243475_81243515 | 0.41 | 3.2e-04 | Click! |
Activity profile of Nfic_Nfib motif
Sorted Z-values of Nfic_Nfib motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfic_Nfib
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_87501548 | 20.64 |
ENSMUST00000068681.12
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr8_+_95498822 | 18.67 |
ENSMUST00000211956.2
ENSMUST00000211947.2 |
Cx3cl1
|
chemokine (C-X3-C motif) ligand 1 |
| chr15_+_54609011 | 18.53 |
ENSMUST00000050027.9
|
Ccn3
|
cellular communication network factor 3 |
| chr2_-_103114105 | 17.36 |
ENSMUST00000111174.8
|
Ehf
|
ets homologous factor |
| chr11_+_78215026 | 16.19 |
ENSMUST00000102478.4
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chrX_-_134968985 | 12.73 |
ENSMUST00000049130.8
|
Bex2
|
brain expressed X-linked 2 |
| chr18_+_35252470 | 12.49 |
ENSMUST00000237154.2
|
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr1_-_75254989 | 12.42 |
ENSMUST00000039534.11
|
Resp18
|
regulated endocrine-specific protein 18 |
| chr19_+_24651362 | 12.07 |
ENSMUST00000057243.6
|
Tmem252
|
transmembrane protein 252 |
| chr6_+_135339929 | 11.80 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr8_+_23247760 | 11.63 |
ENSMUST00000033941.7
|
Plat
|
plasminogen activator, tissue |
| chr11_+_61376257 | 11.51 |
ENSMUST00000064783.10
ENSMUST00000040522.7 |
Mfap4
|
microfibrillar-associated protein 4 |
| chr11_+_3939924 | 11.48 |
ENSMUST00000109981.2
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr2_+_25179903 | 11.46 |
ENSMUST00000028337.7
|
Lrrc26
|
leucine rich repeat containing 26 |
| chr18_+_82572595 | 11.31 |
ENSMUST00000152071.9
ENSMUST00000142850.9 ENSMUST00000080658.12 ENSMUST00000133193.9 ENSMUST00000123251.9 ENSMUST00000153478.9 ENSMUST00000075372.13 ENSMUST00000102812.12 ENSMUST00000062446.15 ENSMUST00000114674.11 ENSMUST00000132369.3 |
Mbp
|
myelin basic protein |
| chr4_+_40920047 | 11.07 |
ENSMUST00000030122.5
|
Spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr14_-_55354392 | 10.29 |
ENSMUST00000022819.13
|
Jph4
|
junctophilin 4 |
| chr10_-_127024641 | 10.15 |
ENSMUST00000218654.2
|
Arhgef25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr6_+_17306334 | 9.88 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr12_+_36042899 | 9.45 |
ENSMUST00000020898.12
|
Agr2
|
anterior gradient 2 |
| chr5_+_9316097 | 9.22 |
ENSMUST00000134991.8
ENSMUST00000069538.14 ENSMUST00000115348.9 |
Elapor2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr18_+_37939442 | 9.20 |
ENSMUST00000076807.7
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3 |
| chr3_-_57202546 | 8.97 |
ENSMUST00000196506.2
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr8_-_95602952 | 8.82 |
ENSMUST00000046461.9
|
Dok4
|
docking protein 4 |
| chr6_+_17306379 | 8.76 |
ENSMUST00000115455.3
|
Cav1
|
caveolin 1, caveolae protein |
| chr18_-_36648850 | 8.63 |
ENSMUST00000025363.7
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr3_-_57202301 | 8.19 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr7_+_40547608 | 8.18 |
ENSMUST00000044705.12
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr6_+_135339543 | 8.08 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr9_+_22012357 | 8.07 |
ENSMUST00000216872.2
|
Cnn1
|
calponin 1 |
| chr2_-_25359752 | 8.04 |
ENSMUST00000114259.3
ENSMUST00000015234.13 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr17_+_28491085 | 7.87 |
ENSMUST00000169040.3
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr9_+_50663171 | 7.86 |
ENSMUST00000214609.2
|
Cryab
|
crystallin, alpha B |
| chr7_-_25516041 | 7.52 |
ENSMUST00000043314.10
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chr12_+_95658987 | 7.48 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr3_+_94840352 | 7.48 |
ENSMUST00000090839.12
|
Selenbp1
|
selenium binding protein 1 |
| chr8_-_65146079 | 7.31 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr10_+_40505985 | 7.28 |
ENSMUST00000019977.8
ENSMUST00000214102.2 ENSMUST00000213503.2 ENSMUST00000213442.2 ENSMUST00000216830.2 |
Ddo
|
D-aspartate oxidase |
| chr2_+_149672760 | 7.18 |
ENSMUST00000109934.2
ENSMUST00000140870.8 |
Syndig1
|
synapse differentiation inducing 1 |
| chr17_+_35188888 | 7.18 |
ENSMUST00000173680.2
|
Gm20481
|
predicted gene 20481 |
| chr4_+_137408975 | 7.17 |
ENSMUST00000047243.12
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr7_+_111825063 | 7.05 |
ENSMUST00000050149.12
|
Mical2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr16_+_34605282 | 7.01 |
ENSMUST00000023538.9
|
Mylk
|
myosin, light polypeptide kinase |
| chr18_-_64688271 | 6.90 |
ENSMUST00000235459.2
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr4_-_119515978 | 6.84 |
ENSMUST00000106309.9
ENSMUST00000044426.8 |
Guca2b
|
guanylate cyclase activator 2b (retina) |
| chr6_+_21986445 | 6.80 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr17_-_27947863 | 6.78 |
ENSMUST00000167489.2
ENSMUST00000138970.3 ENSMUST00000025054.10 ENSMUST00000114870.9 |
Spdef
|
SAM pointed domain containing ets transcription factor |
| chr17_+_43671314 | 6.75 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr17_+_37253802 | 6.67 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr15_-_75963446 | 6.65 |
ENSMUST00000228366.3
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr6_+_17306414 | 6.54 |
ENSMUST00000150901.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr5_+_35971767 | 6.50 |
ENSMUST00000114203.8
ENSMUST00000150146.2 |
Ablim2
|
actin-binding LIM protein 2 |
| chr6_+_29433247 | 6.48 |
ENSMUST00000101617.9
ENSMUST00000065090.8 |
Flnc
|
filamin C, gamma |
| chr9_+_44045859 | 6.46 |
ENSMUST00000034650.15
ENSMUST00000098852.3 ENSMUST00000216002.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr4_-_155103837 | 6.37 |
ENSMUST00000126098.2
ENSMUST00000176194.8 |
Plch2
|
phospholipase C, eta 2 |
| chrX_+_149829131 | 6.34 |
ENSMUST00000112685.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr2_+_156317416 | 6.23 |
ENSMUST00000029155.16
|
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr14_+_103750814 | 6.22 |
ENSMUST00000095576.5
|
Scel
|
sciellin |
| chr15_-_73114855 | 6.19 |
ENSMUST00000227686.2
|
Ptk2
|
PTK2 protein tyrosine kinase 2 |
| chr14_+_70768257 | 6.16 |
ENSMUST00000047331.8
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr9_-_60557076 | 6.07 |
ENSMUST00000053171.14
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr2_-_164246526 | 6.02 |
ENSMUST00000109359.8
ENSMUST00000109358.8 ENSMUST00000103103.10 |
Matn4
|
matrilin 4 |
| chr15_+_102011352 | 6.01 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr5_-_135107078 | 5.90 |
ENSMUST00000067935.11
ENSMUST00000076203.3 |
Vps37d
|
vacuolar protein sorting 37D |
| chr2_-_25360043 | 5.87 |
ENSMUST00000114251.8
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr6_-_115229128 | 5.87 |
ENSMUST00000032462.9
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr15_+_102011415 | 5.83 |
ENSMUST00000046144.10
|
Tns2
|
tensin 2 |
| chr14_+_103750763 | 5.76 |
ENSMUST00000227322.2
|
Scel
|
sciellin |
| chr11_+_78213791 | 5.74 |
ENSMUST00000017534.15
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr2_+_149672708 | 5.61 |
ENSMUST00000109935.8
|
Syndig1
|
synapse differentiation inducing 1 |
| chr14_+_70768289 | 5.45 |
ENSMUST00000226548.2
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr6_-_115228800 | 5.40 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chrX_+_165021919 | 5.40 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr11_+_66847446 | 5.27 |
ENSMUST00000211300.2
ENSMUST00000150220.2 |
Tmem238l
|
transmembrane protein 238 like |
| chr3_-_72964276 | 5.19 |
ENSMUST00000192477.2
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr3_-_85653573 | 5.18 |
ENSMUST00000118408.8
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr3_+_62245765 | 5.17 |
ENSMUST00000079300.13
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr9_-_107483855 | 5.16 |
ENSMUST00000073448.12
ENSMUST00000194606.2 ENSMUST00000195662.6 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr5_+_35971697 | 5.11 |
ENSMUST00000130233.8
|
Ablim2
|
actin-binding LIM protein 2 |
| chr9_+_50662625 | 5.05 |
ENSMUST00000217475.2
|
Cryab
|
crystallin, alpha B |
| chr2_-_26012751 | 4.98 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr11_+_96822213 | 4.97 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr9_-_56068282 | 4.87 |
ENSMUST00000034876.10
|
Tspan3
|
tetraspanin 3 |
| chr3_-_107129038 | 4.87 |
ENSMUST00000029504.9
|
Cym
|
chymosin |
| chr9_-_99592116 | 4.84 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr5_-_77262968 | 4.73 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr1_+_133173826 | 4.70 |
ENSMUST00000105082.9
ENSMUST00000038295.15 |
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr15_+_81686622 | 4.68 |
ENSMUST00000109553.10
|
Tef
|
thyrotroph embryonic factor |
| chr9_-_99592058 | 4.66 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr5_-_139798992 | 4.60 |
ENSMUST00000110832.8
|
Tmem184a
|
transmembrane protein 184a |
| chr12_+_108376884 | 4.58 |
ENSMUST00000109857.8
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr3_-_63758672 | 4.56 |
ENSMUST00000162269.9
ENSMUST00000159676.9 ENSMUST00000175947.8 |
Plch1
|
phospholipase C, eta 1 |
| chr2_-_118593087 | 4.53 |
ENSMUST00000059997.9
|
Ccdc9b
|
coiled-coil domain containing 9B |
| chr11_+_3438274 | 4.53 |
ENSMUST00000064265.13
|
Pla2g3
|
phospholipase A2, group III |
| chr12_+_108376801 | 4.52 |
ENSMUST00000054955.14
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr19_+_4644365 | 4.51 |
ENSMUST00000113825.4
|
Pcx
|
pyruvate carboxylase |
| chr15_+_34306812 | 4.50 |
ENSMUST00000226766.2
ENSMUST00000163455.9 ENSMUST00000022947.7 ENSMUST00000228570.2 ENSMUST00000227759.2 |
Matn2
|
matrilin 2 |
| chr3_+_90434160 | 4.49 |
ENSMUST00000199538.5
ENSMUST00000164481.7 ENSMUST00000167598.6 |
S100a14
|
S100 calcium binding protein A14 |
| chr7_+_34885782 | 4.46 |
ENSMUST00000135452.8
ENSMUST00000001854.12 |
Slc7a10
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 10 |
| chr16_-_30369378 | 4.45 |
ENSMUST00000140402.8
|
Tmem44
|
transmembrane protein 44 |
| chr3_-_63872079 | 4.44 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr2_+_149672814 | 4.41 |
ENSMUST00000137280.2
ENSMUST00000149705.2 |
Syndig1
|
synapse differentiation inducing 1 |
| chr2_+_165345707 | 4.39 |
ENSMUST00000029196.5
|
Slc2a10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chrX_+_165021897 | 4.34 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chr5_+_130398261 | 4.30 |
ENSMUST00000086029.10
|
Caln1
|
calneuron 1 |
| chr19_-_61129211 | 4.29 |
ENSMUST00000143264.8
ENSMUST00000205854.2 |
Zfp950
|
zinc finger protein 950 |
| chr4_+_120018403 | 4.29 |
ENSMUST00000030384.5
|
Edn2
|
endothelin 2 |
| chrX_-_51702790 | 4.27 |
ENSMUST00000069360.14
|
Gpc3
|
glypican 3 |
| chr1_+_165130192 | 4.24 |
ENSMUST00000111450.3
|
Gpr161
|
G protein-coupled receptor 161 |
| chr11_-_97040858 | 4.23 |
ENSMUST00000118375.8
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr12_+_74044435 | 4.22 |
ENSMUST00000221220.2
|
Syt16
|
synaptotagmin XVI |
| chr13_-_98951627 | 4.21 |
ENSMUST00000224992.2
ENSMUST00000225840.2 |
Fcho2
|
FCH domain only 2 |
| chr11_-_58504307 | 4.19 |
ENSMUST00000048801.8
|
Lypd8l
|
LY6/PLAUR domain containing 8 like |
| chr11_-_76359222 | 4.16 |
ENSMUST00000065028.14
|
Abr
|
active BCR-related gene |
| chr4_+_43506966 | 4.13 |
ENSMUST00000030183.10
|
Car9
|
carbonic anhydrase 9 |
| chr19_-_11796085 | 4.09 |
ENSMUST00000211047.2
ENSMUST00000075304.14 ENSMUST00000211641.2 |
Stx3
|
syntaxin 3 |
| chr2_-_179867605 | 4.09 |
ENSMUST00000015791.6
|
Lama5
|
laminin, alpha 5 |
| chr4_+_129941633 | 4.07 |
ENSMUST00000044565.15
ENSMUST00000132251.2 |
Col16a1
|
collagen, type XVI, alpha 1 |
| chrX_+_135171002 | 3.98 |
ENSMUST00000178632.8
ENSMUST00000053540.11 |
Bex3
|
brain expressed X-linked 3 |
| chr5_-_35828857 | 3.94 |
ENSMUST00000129459.2
ENSMUST00000137935.2 |
Htra3
|
HtrA serine peptidase 3 |
| chr1_-_162687254 | 3.93 |
ENSMUST00000131058.8
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr4_-_155095441 | 3.92 |
ENSMUST00000105631.9
ENSMUST00000139976.9 ENSMUST00000145662.9 |
Plch2
|
phospholipase C, eta 2 |
| chr1_-_72914036 | 3.91 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr10_-_81127057 | 3.89 |
ENSMUST00000045744.7
|
Tjp3
|
tight junction protein 3 |
| chr11_-_86964881 | 3.88 |
ENSMUST00000020804.8
|
Gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr19_-_61129199 | 3.87 |
ENSMUST00000180544.3
|
Zfp950
|
zinc finger protein 950 |
| chr13_-_3968157 | 3.85 |
ENSMUST00000223258.2
ENSMUST00000091853.12 |
Net1
|
neuroepithelial cell transforming gene 1 |
| chr1_-_190915441 | 3.82 |
ENSMUST00000027941.14
|
Atf3
|
activating transcription factor 3 |
| chr3_-_89152320 | 3.79 |
ENSMUST00000107464.8
ENSMUST00000090924.13 |
Trim46
|
tripartite motif-containing 46 |
| chr8_-_106223502 | 3.79 |
ENSMUST00000212303.2
|
Zdhhc1
|
zinc finger, DHHC domain containing 1 |
| chr17_-_31383976 | 3.78 |
ENSMUST00000235870.2
|
Tff1
|
trefoil factor 1 |
| chrX_+_136552469 | 3.77 |
ENSMUST00000075471.4
|
Il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr1_-_162305573 | 3.75 |
ENSMUST00000086074.12
ENSMUST00000070330.14 |
Dnm3
|
dynamin 3 |
| chr4_-_129121676 | 3.75 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr16_+_58228806 | 3.75 |
ENSMUST00000046663.8
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr3_+_94600863 | 3.73 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr5_+_150119860 | 3.72 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr7_-_118594365 | 3.69 |
ENSMUST00000008878.10
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B |
| chr7_+_130633776 | 3.67 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr4_-_57916283 | 3.67 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr11_-_100012384 | 3.67 |
ENSMUST00000007275.3
|
Krt13
|
keratin 13 |
| chr5_-_137919873 | 3.65 |
ENSMUST00000031741.8
|
Cyp3a13
|
cytochrome P450, family 3, subfamily a, polypeptide 13 |
| chrX_-_51702813 | 3.63 |
ENSMUST00000114857.2
|
Gpc3
|
glypican 3 |
| chr3_+_27371206 | 3.62 |
ENSMUST00000174840.2
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr11_+_98727611 | 3.61 |
ENSMUST00000107479.3
|
Rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chrX_+_150961960 | 3.61 |
ENSMUST00000096275.5
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr1_-_79838897 | 3.61 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr5_-_53864874 | 3.58 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr11_+_28803188 | 3.58 |
ENSMUST00000020759.12
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chr19_+_4644425 | 3.57 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr16_+_33712305 | 3.56 |
ENSMUST00000232262.2
|
Itgb5
|
integrin beta 5 |
| chr3_-_107667499 | 3.52 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr2_+_118493713 | 3.50 |
ENSMUST00000099557.10
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr7_-_105131407 | 3.49 |
ENSMUST00000047040.4
|
Cavin3
|
caveolae associated 3 |
| chr2_+_177834868 | 3.49 |
ENSMUST00000103065.2
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr17_+_71980249 | 3.49 |
ENSMUST00000097284.10
|
Togaram2
|
TOG array regulator of axonemal microtubules 2 |
| chr1_+_172328768 | 3.46 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chrX_+_135171049 | 3.46 |
ENSMUST00000113112.2
|
Bex3
|
brain expressed X-linked 3 |
| chr7_-_28297565 | 3.45 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chr13_-_6686686 | 3.43 |
ENSMUST00000136585.2
|
Pfkp
|
phosphofructokinase, platelet |
| chr4_-_115504907 | 3.42 |
ENSMUST00000102707.10
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr4_+_140966810 | 3.40 |
ENSMUST00000141834.9
|
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr2_+_84880776 | 3.38 |
ENSMUST00000111605.9
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr3_-_89153135 | 3.36 |
ENSMUST00000041022.15
|
Trim46
|
tripartite motif-containing 46 |
| chr3_+_96508400 | 3.34 |
ENSMUST00000062058.5
|
Lix1l
|
Lix1-like |
| chr13_-_63006176 | 3.31 |
ENSMUST00000021907.9
|
Fbp2
|
fructose bisphosphatase 2 |
| chr12_+_16703383 | 3.30 |
ENSMUST00000221596.2
ENSMUST00000111064.3 ENSMUST00000220892.2 |
Ntsr2
|
neurotensin receptor 2 |
| chr16_-_17394495 | 3.29 |
ENSMUST00000231645.2
ENSMUST00000232226.2 ENSMUST00000232336.2 ENSMUST00000232385.2 ENSMUST00000231615.2 ENSMUST00000231283.2 ENSMUST00000172164.10 |
Slc7a4
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 4 |
| chr10_-_18619658 | 3.25 |
ENSMUST00000215836.2
|
Arfgef3
|
ARFGEF family member 3 |
| chr3_-_94566107 | 3.20 |
ENSMUST00000196655.5
ENSMUST00000200407.2 ENSMUST00000006123.11 ENSMUST00000196733.5 |
Tuft1
|
tuftelin 1 |
| chr8_+_13485451 | 3.20 |
ENSMUST00000167505.3
ENSMUST00000167071.9 |
Tmem255b
|
transmembrane protein 255B |
| chr19_-_6134703 | 3.19 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1 |
| chr11_+_98337655 | 3.18 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr19_-_34579356 | 3.17 |
ENSMUST00000168254.3
|
Ifit1bl1
|
interferon induced protein with tetratricpeptide repeats 1B like 1 |
| chr1_+_75376714 | 3.16 |
ENSMUST00000113589.8
|
Speg
|
SPEG complex locus |
| chr3_+_13538266 | 3.15 |
ENSMUST00000211860.2
|
Ralyl
|
RALY RNA binding protein-like |
| chr9_-_21710029 | 3.14 |
ENSMUST00000034717.7
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr19_-_5148506 | 3.14 |
ENSMUST00000025805.8
|
Cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr4_-_9643636 | 3.13 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr9_+_44684324 | 3.13 |
ENSMUST00000214854.3
ENSMUST00000125877.8 |
Ift46
|
intraflagellar transport 46 |
| chr2_+_158354716 | 3.10 |
ENSMUST00000165398.2
ENSMUST00000099133.10 |
Arhgap40
|
Rho GTPase activating protein 40 |
| chr18_+_61096660 | 3.09 |
ENSMUST00000039904.7
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr17_+_37253916 | 3.08 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr8_-_37200051 | 3.08 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr2_+_164327988 | 3.06 |
ENSMUST00000109350.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr3_-_107838895 | 3.06 |
ENSMUST00000133947.9
ENSMUST00000124215.2 ENSMUST00000106688.8 ENSMUST00000106687.9 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr2_+_83642910 | 3.05 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chrX_-_47123719 | 3.04 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr17_+_31515163 | 3.02 |
ENSMUST00000235972.2
ENSMUST00000165149.3 ENSMUST00000236251.2 |
Slc37a1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
| chr1_-_162687369 | 3.02 |
ENSMUST00000193078.6
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr17_+_32877851 | 3.00 |
ENSMUST00000235086.2
|
Cyp4f40
|
cytochrome P450, family 4, subfamily f, polypeptide 40 |
| chr2_+_152596075 | 3.00 |
ENSMUST00000010020.12
|
Cox4i2
|
cytochrome c oxidase subunit 4I2 |
| chr6_+_54658609 | 2.99 |
ENSMUST00000190641.7
ENSMUST00000187701.2 |
Mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr7_-_19530714 | 2.98 |
ENSMUST00000108449.9
ENSMUST00000043822.8 |
Cblc
|
Casitas B-lineage lymphoma c |
| chr7_+_81984940 | 2.96 |
ENSMUST00000173287.8
|
Adamtsl3
|
ADAMTS-like 3 |
| chr9_-_15212745 | 2.96 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr11_-_3489228 | 2.95 |
ENSMUST00000075118.10
ENSMUST00000136243.2 ENSMUST00000020721.15 ENSMUST00000170588.8 |
Smtn
|
smoothelin |
| chr18_+_61096597 | 2.92 |
ENSMUST00000115295.9
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr9_+_88209250 | 2.91 |
ENSMUST00000034992.8
|
Nt5e
|
5' nucleotidase, ecto |
| chr3_-_94693740 | 2.91 |
ENSMUST00000153263.9
ENSMUST00000107272.7 ENSMUST00000155485.4 |
Cgn
|
cingulin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 25.2 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 3.7 | 18.7 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 3.1 | 9.4 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 2.9 | 8.6 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 2.8 | 11.3 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 2.4 | 7.3 | GO:0030070 | insulin processing(GO:0030070) |
| 2.4 | 7.3 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 2.4 | 7.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 2.3 | 13.9 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 2.1 | 12.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 2.0 | 9.9 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 1.8 | 7.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 1.7 | 5.0 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 1.6 | 8.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.6 | 7.9 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 1.6 | 28.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.5 | 24.4 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 1.5 | 4.5 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 1.5 | 4.5 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 1.5 | 4.5 | GO:0015825 | L-serine transport(GO:0015825) |
| 1.4 | 4.3 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.4 | 4.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.4 | 7.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.4 | 9.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 1.3 | 11.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.3 | 3.8 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 1.2 | 4.9 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.2 | 9.7 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 3.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 1.2 | 7.2 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 1.2 | 12.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.2 | 3.5 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 1.2 | 3.5 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 1.2 | 11.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.2 | 3.5 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 1.1 | 12.6 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 1.1 | 3.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.1 | 2.1 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 1.0 | 3.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.0 | 6.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 1.0 | 11.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.0 | 3.0 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.0 | 3.0 | GO:1901662 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 1.0 | 3.9 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.9 | 3.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.9 | 9.0 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.9 | 6.9 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.9 | 1.7 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.8 | 8.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.8 | 19.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.8 | 3.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.8 | 17.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.8 | 0.8 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.8 | 6.2 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.8 | 4.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.8 | 4.5 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.7 | 4.4 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.7 | 2.9 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.7 | 0.7 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.7 | 14.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.7 | 2.7 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.7 | 2.0 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.7 | 3.3 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.6 | 3.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.6 | 1.8 | GO:0061622 | UDP-glucose metabolic process(GO:0006011) glycolytic process through glucose-1-phosphate(GO:0061622) glycolytic process from galactose(GO:0061623) |
| 0.6 | 3.0 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.6 | 3.6 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.6 | 5.9 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.5 | 7.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.5 | 2.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.5 | 6.8 | GO:0060480 | positive regulation of cell fate commitment(GO:0010455) lung goblet cell differentiation(GO:0060480) intestinal epithelial cell development(GO:0060576) |
| 0.5 | 10.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.5 | 2.0 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.5 | 2.5 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.5 | 2.4 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.5 | 2.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.4 | 1.8 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.4 | 4.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.4 | 6.0 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.4 | 3.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 1.3 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 1.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 2.1 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.4 | 4.1 | GO:0098881 | synaptic vesicle docking(GO:0016081) exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.4 | 7.7 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.4 | 4.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 1.6 | GO:0090346 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.4 | 2.4 | GO:0009624 | response to nematode(GO:0009624) |
| 0.4 | 2.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 9.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 2.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 2.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.4 | 4.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.4 | 4.5 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.4 | 0.7 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 | 2.2 | GO:0061718 | NADH regeneration(GO:0006735) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 1.1 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.4 | 1.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.3 | 1.4 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.3 | 1.4 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.3 | 7.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.3 | 2.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.3 | 1.0 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.3 | 6.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 1.0 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.3 | 2.4 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.3 | 0.9 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.3 | 0.9 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.3 | 2.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.3 | 2.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.3 | 3.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.3 | 10.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 3.9 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.3 | 1.9 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.3 | 1.9 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.3 | 1.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 1.3 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.3 | 5.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.3 | 2.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.3 | 10.3 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.3 | 3.9 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 4.9 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.3 | 0.8 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.2 | 1.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 0.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.2 | 8.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.2 | 4.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.2 | 1.4 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 1.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 1.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 4.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 1.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.9 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 6.4 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 3.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 0.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.2 | 3.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 1.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.2 | 0.9 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.2 | 0.9 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.2 | 0.7 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 6.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 2.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 6.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 2.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 0.5 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.2 | 4.5 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.2 | 4.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 0.3 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 0.8 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.2 | 0.5 | GO:0014862 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 0.2 | 2.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 1.8 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 6.2 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.1 | 6.8 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 4.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 3.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 3.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 3.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 3.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 1.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 1.4 | GO:1904415 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.1 | 1.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 13.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.5 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.1 | 1.7 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.1 | 0.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.9 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 1.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.7 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 1.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.1 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.1 | 2.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.1 | 3.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.9 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.1 | 6.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 5.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 3.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.4 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.4 | GO:1902302 | regulation of potassium ion export(GO:1902302) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 4.1 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.1 | 4.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 2.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 5.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 1.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.8 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 0.7 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 4.6 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.1 | 5.5 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.1 | 1.5 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.3 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 3.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 4.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.3 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) |
| 0.1 | 8.9 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.1 | 4.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 5.8 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 2.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 6.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 5.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 11.7 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.1 | 1.4 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 3.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 4.0 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 0.3 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.1 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.7 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 1.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.8 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 | 6.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 13.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 4.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 1.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.2 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.0 | 3.1 | GO:0032414 | positive regulation of ion transmembrane transporter activity(GO:0032414) |
| 0.0 | 5.1 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 2.5 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 1.9 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 1.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 2.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 2.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 1.0 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 3.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 2.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.2 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 1.4 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.3 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 2.3 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 4.9 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 2.6 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 1.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.6 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 2.3 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 8.9 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 1.2 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 5.8 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 3.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 1.3 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 2.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 2.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 13.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.8 | 7.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.8 | 12.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.5 | 6.0 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 1.5 | 25.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.4 | 11.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.4 | 4.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 1.3 | 3.8 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 1.3 | 3.8 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 1.3 | 3.8 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 1.2 | 4.8 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 1.0 | 4.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.9 | 10.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.9 | 13.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.9 | 3.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.8 | 10.2 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.7 | 3.6 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.7 | 4.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.7 | 12.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.6 | 2.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.5 | 2.7 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.4 | 10.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.4 | 7.4 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.4 | 5.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 4.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.4 | 2.4 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.4 | 24.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.4 | 3.9 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 6.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 4.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 3.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 0.8 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.3 | 3.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 1.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 7.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 8.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 3.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 1.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 0.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 1.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 2.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 8.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 15.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 4.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.8 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 4.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 6.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 3.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 1.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.5 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 2.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 2.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 4.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 4.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 6.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.6 | GO:0032839 | autophagosome membrane(GO:0000421) dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 9.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 9.4 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 9.5 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 1.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 5.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 28.6 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.1 | 3.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 9.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 3.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 8.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 9.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 9.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 7.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 3.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 2.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 5.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 2.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 9.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 6.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 25.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 7.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 4.7 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 8.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 3.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 8.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 7.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 2.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 7.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 5.1 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 0.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.2 | GO:1990204 | oxidoreductase complex(GO:1990204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 25.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 3.5 | 13.9 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 3.0 | 11.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 2.6 | 7.9 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 2.2 | 21.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.0 | 8.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.8 | 7.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 1.5 | 4.6 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 1.5 | 4.5 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 1.3 | 6.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 1.3 | 3.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 1.2 | 3.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.2 | 3.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.1 | 6.8 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 1.1 | 4.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.9 | 3.8 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.9 | 13.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.9 | 3.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 6.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.9 | 7.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.8 | 5.9 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.8 | 4.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.8 | 3.8 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.8 | 3.0 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.7 | 2.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.7 | 4.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 6.9 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.7 | 7.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.6 | 11.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.6 | 2.5 | GO:0051381 | histamine binding(GO:0051381) |
| 0.6 | 2.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.6 | 2.3 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.6 | 14.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.5 | 2.7 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.5 | 11.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 2.4 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.5 | 3.4 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.5 | 18.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 2.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.4 | 12.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.4 | 2.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 1.3 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.4 | 9.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 1.6 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.4 | 1.6 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.4 | 7.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.4 | 3.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 4.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 7.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 1.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 4.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 1.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 1.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.3 | 4.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.0 | GO:0070025 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitric oxide binding(GO:0070026) |
| 0.3 | 4.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 10.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 1.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.3 | 1.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.3 | 5.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 2.2 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 6.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 7.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 41.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.3 | 4.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 1.8 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.3 | 11.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.3 | 2.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 0.8 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.3 | 2.5 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.2 | 7.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 10.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.7 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 1.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 1.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 4.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 0.7 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.2 | 4.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 2.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 11.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 0.6 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.2 | 4.5 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.2 | 1.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 1.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 3.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 3.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 1.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 6.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 6.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 1.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 2.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 5.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 2.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 4.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 1.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 4.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 2.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 0.5 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.2 | 8.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 4.1 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 6.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 10.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 4.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 4.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 8.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 13.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 14.1 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 10.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 22.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.9 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 2.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 4.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 4.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 0.8 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 2.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.7 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 3.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 4.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 2.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.3 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 4.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 4.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 1.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 0.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 1.1 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 0.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 1.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.7 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 1.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 6.9 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 2.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 3.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 9.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.5 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.1 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.3 | GO:0047555 | cGMP binding(GO:0030553) 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 3.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.9 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.3 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 22.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 2.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 12.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 2.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.1 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 4.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 4.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 2.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 2.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.5 | GO:0016836 | hydro-lyase activity(GO:0016836) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 25.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.5 | 37.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.5 | 18.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.3 | 9.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 20.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 5.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 4.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 3.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 12.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 12.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 42.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 4.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 6.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 6.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 27.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 7.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 26.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 7.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 3.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 9.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 23.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.5 | 25.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 5.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 5.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 6.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 18.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 31.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 8.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 14.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 7.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 4.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 10.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 1.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 3.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 5.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 12.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 6.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 7.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 9.5 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.2 | 7.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 11.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 7.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 14.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 4.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 3.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 3.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 4.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 3.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 3.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 31.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 3.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 4.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 10.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 1.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 4.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 5.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 4.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 1.8 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |