Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
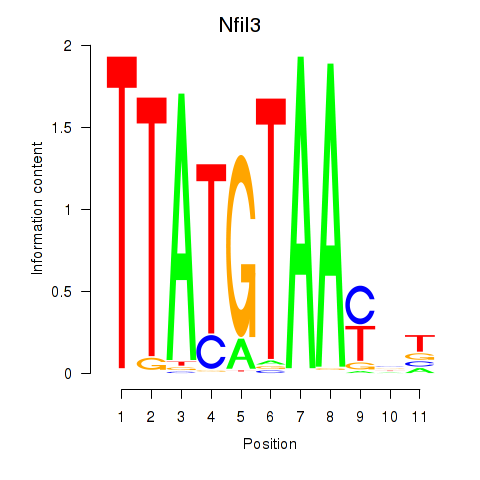
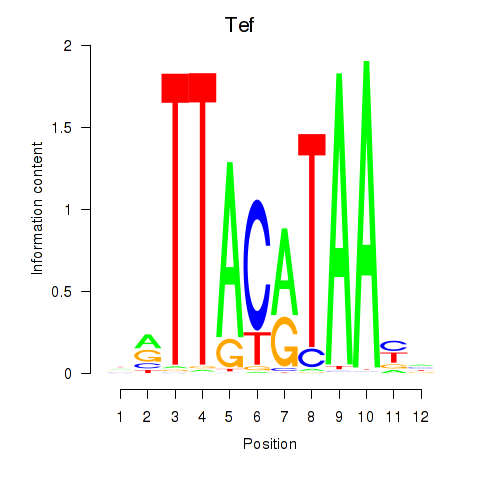
Results for Nfil3_Tef
Z-value: 2.52
Transcription factors associated with Nfil3_Tef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfil3
|
ENSMUSG00000056749.8 | Nfil3 |
|
Tef
|
ENSMUSG00000022389.16 | Tef |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tef | mm39_v1_chr15_+_81686622_81686779 | 0.67 | 1.3e-10 | Click! |
| Nfil3 | mm39_v1_chr13_-_53135064_53135114 | 0.38 | 1.1e-03 | Click! |
Activity profile of Nfil3_Tef motif
Sorted Z-values of Nfil3_Tef motif
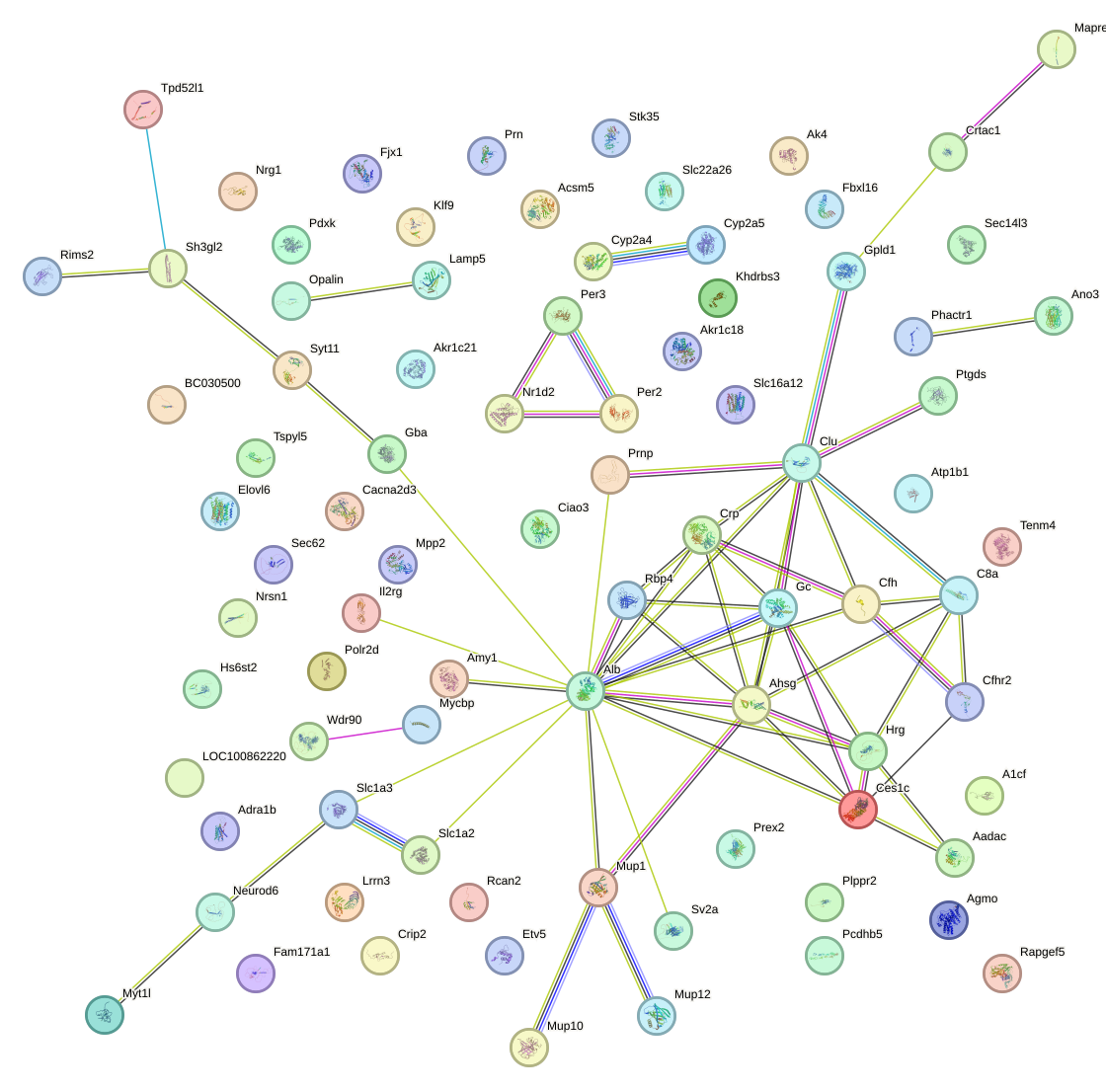
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfil3_Tef
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_26006594 | 29.79 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr14_+_40827317 | 28.21 |
ENSMUST00000047286.7
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr6_-_113911640 | 27.49 |
ENSMUST00000101044.9
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr14_+_40826970 | 26.17 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr14_+_66208498 | 25.57 |
ENSMUST00000128539.8
|
Clu
|
clusterin |
| chr13_-_25454058 | 24.65 |
ENSMUST00000057866.13
|
Nrsn1
|
neurensin 1 |
| chr6_-_55658242 | 23.78 |
ENSMUST00000044767.10
|
Neurod6
|
neurogenic differentiation 6 |
| chr14_+_40827108 | 23.08 |
ENSMUST00000224514.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr14_+_66208253 | 22.93 |
ENSMUST00000138191.8
|
Clu
|
clusterin |
| chr5_+_90608751 | 22.77 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr19_-_38113696 | 22.53 |
ENSMUST00000025951.14
ENSMUST00000237287.2 |
Rbp4
|
retinol binding protein 4, plasma |
| chr19_-_41065544 | 22.36 |
ENSMUST00000087176.8
|
Opalin
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr14_+_66208059 | 22.19 |
ENSMUST00000127387.8
|
Clu
|
clusterin |
| chr7_+_26534730 | 22.10 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr14_+_66208613 | 20.77 |
ENSMUST00000144619.2
|
Clu
|
clusterin |
| chr12_-_81014849 | 19.93 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr16_+_22769822 | 18.87 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr14_+_60615128 | 18.71 |
ENSMUST00000022561.9
|
Amer2
|
APC membrane recruitment 2 |
| chr11_-_43792013 | 18.17 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr2_-_66271097 | 18.01 |
ENSMUST00000112371.9
ENSMUST00000138910.3 |
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr2_+_102536701 | 17.96 |
ENSMUST00000123759.8
ENSMUST00000005220.11 ENSMUST00000111212.8 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr4_+_101276474 | 17.27 |
ENSMUST00000102780.8
ENSMUST00000106946.8 ENSMUST00000106945.8 |
Ak4
|
adenylate kinase 4 |
| chr3_-_113368407 | 17.26 |
ENSMUST00000106540.8
|
Amy1
|
amylase 1, salivary |
| chr12_-_81014755 | 17.05 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr4_+_101276893 | 16.78 |
ENSMUST00000131397.2
ENSMUST00000133055.8 |
Ak4
|
adenylate kinase 4 |
| chr16_+_22769844 | 15.84 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr2_-_25359752 | 15.07 |
ENSMUST00000114259.3
ENSMUST00000015234.13 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr1_-_91386976 | 14.96 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2 |
| chr4_+_85123654 | 14.51 |
ENSMUST00000030212.15
ENSMUST00000107189.8 ENSMUST00000107184.8 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr8_-_93857899 | 13.47 |
ENSMUST00000034189.17
|
Ces1c
|
carboxylesterase 1C |
| chr17_+_26028059 | 13.47 |
ENSMUST00000045692.9
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr4_+_101365052 | 13.43 |
ENSMUST00000038207.12
|
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr7_+_143729250 | 13.38 |
ENSMUST00000105900.9
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr16_+_22710785 | 13.22 |
ENSMUST00000023583.7
ENSMUST00000232098.2 |
Ahsg
|
alpha-2-HS-glycoprotein |
| chr2_-_25360043 | 13.17 |
ENSMUST00000114251.8
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr4_+_101365144 | 12.86 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr2_+_135899847 | 11.75 |
ENSMUST00000057503.7
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr17_+_44112679 | 11.18 |
ENSMUST00000229744.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr5_-_89605622 | 11.07 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr19_-_34724689 | 11.02 |
ENSMUST00000009522.4
|
Slc16a12
|
solute carrier family 16 (monocarboxylic acid transporters), member 12 |
| chr1_-_58625350 | 10.83 |
ENSMUST00000038372.14
ENSMUST00000097724.10 |
Fam126b
|
family with sequence similarity 126, member B |
| chr2_+_131751848 | 10.73 |
ENSMUST00000091288.13
ENSMUST00000124100.8 ENSMUST00000136783.2 |
Prnp
Prn
|
prion protein prion protein readthrough transcript |
| chrX_-_50770580 | 10.29 |
ENSMUST00000088172.12
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chrX_-_50770733 | 10.17 |
ENSMUST00000114871.2
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr3_+_129326285 | 9.95 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr2_-_110781268 | 9.87 |
ENSMUST00000099623.10
|
Ano3
|
anoctamin 3 |
| chr2_-_102282844 | 9.83 |
ENSMUST00000099678.5
|
Fjx1
|
four jointed box 1 |
| chr11_+_78181667 | 9.81 |
ENSMUST00000046361.5
ENSMUST00000238934.2 |
Rskr
|
ribosomal protein S6 kinase related |
| chr15_+_68800261 | 9.77 |
ENSMUST00000022954.7
|
Khdrbs3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr12_+_117621644 | 9.63 |
ENSMUST00000222105.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_+_85123358 | 9.43 |
ENSMUST00000107188.10
|
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr11_-_101979297 | 9.40 |
ENSMUST00000017458.11
|
Mpp2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr4_-_60538151 | 9.29 |
ENSMUST00000098047.3
|
Mup10
|
major urinary protein 10 |
| chr10_-_31321793 | 9.19 |
ENSMUST00000213639.2
ENSMUST00000215515.2 ENSMUST00000214644.2 ENSMUST00000213528.2 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr1_+_172525613 | 9.15 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr3_+_96088467 | 8.99 |
ENSMUST00000035371.9
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr6_+_133082202 | 8.74 |
ENSMUST00000191462.2
|
Smim10l1
|
small integral membrane protein 10 like 1 |
| chr6_+_106095726 | 8.64 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chr18_+_23937019 | 8.61 |
ENSMUST00000025127.5
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_-_164281344 | 8.56 |
ENSMUST00000193367.2
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr4_-_104733580 | 8.56 |
ENSMUST00000064873.9
ENSMUST00000106808.10 ENSMUST00000048947.15 |
C8a
|
complement component 8, alpha polypeptide |
| chr7_+_95860863 | 8.53 |
ENSMUST00000107165.8
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr4_-_41774097 | 8.45 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr14_+_60615690 | 8.38 |
ENSMUST00000239079.2
|
Amer2
|
APC membrane recruitment 2 |
| chr9_+_21848282 | 8.31 |
ENSMUST00000046371.13
|
Plppr2
|
phospholipid phosphatase related 2 |
| chr3_+_141170941 | 8.28 |
ENSMUST00000106236.9
|
Unc5c
|
unc-5 netrin receptor C |
| chr7_+_119125443 | 8.05 |
ENSMUST00000207440.2
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr15_-_8739893 | 7.95 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr1_+_11063678 | 7.71 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr15_-_33687986 | 7.70 |
ENSMUST00000042021.5
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr13_+_76727787 | 7.69 |
ENSMUST00000126960.8
ENSMUST00000109583.9 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr7_+_119125426 | 7.69 |
ENSMUST00000066465.3
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr7_+_119125546 | 7.43 |
ENSMUST00000207387.2
ENSMUST00000207813.2 |
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr3_-_88669551 | 7.22 |
ENSMUST00000183267.2
|
Syt11
|
synaptotagmin XI |
| chr2_+_129642371 | 7.14 |
ENSMUST00000165413.9
ENSMUST00000166282.3 |
Stk35
|
serine/threonine kinase 35 |
| chr11_+_4014841 | 7.11 |
ENSMUST00000068322.7
|
Sec14l3
|
SEC14-like lipid binding 3 |
| chr1_-_140111138 | 6.88 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr13_+_43019718 | 6.88 |
ENSMUST00000131942.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr18_+_37453427 | 6.87 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr3_+_30847024 | 6.81 |
ENSMUST00000029256.9
|
Sec62
|
SEC62 homolog (S. cerevisiae) |
| chr3_+_59939175 | 6.75 |
ENSMUST00000029325.5
|
Aadac
|
arylacetamide deacetylase |
| chr14_-_28691423 | 6.71 |
ENSMUST00000225985.2
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr16_-_22258320 | 6.65 |
ENSMUST00000170393.2
|
Etv5
|
ets variant 5 |
| chr15_-_8740218 | 6.63 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr10_-_78300802 | 6.62 |
ENSMUST00000041616.15
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr14_+_4230569 | 6.55 |
ENSMUST00000090543.6
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr13_+_4624074 | 6.54 |
ENSMUST00000021628.4
|
Akr1c21
|
aldo-keto reductase family 1, member C21 |
| chr1_-_140111018 | 6.34 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr8_+_59365291 | 6.24 |
ENSMUST00000160055.2
|
BC030500
|
cDNA sequence BC030500 |
| chr19_+_31846154 | 6.21 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr19_-_42420216 | 6.21 |
ENSMUST00000048630.8
ENSMUST00000238290.2 |
Crtac1
|
cartilage acidic protein 1 |
| chr2_+_3115250 | 6.15 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr13_+_25127127 | 6.07 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr19_+_23118545 | 6.07 |
ENSMUST00000036884.3
|
Klf9
|
Kruppel-like factor 9 |
| chr19_-_7779943 | 6.06 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr2_-_66240408 | 6.05 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr15_+_39255185 | 6.04 |
ENSMUST00000228839.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr13_-_4200627 | 5.94 |
ENSMUST00000110704.9
ENSMUST00000021635.9 |
Akr1c18
|
aldo-keto reductase family 1, member C18 |
| chr3_+_141171347 | 5.90 |
ENSMUST00000130636.8
|
Unc5c
|
unc-5 netrin receptor C |
| chr10_-_31321938 | 5.81 |
ENSMUST00000000305.7
|
Tpd52l1
|
tumor protein D52-like 1 |
| chr19_-_7780025 | 5.77 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr12_+_29584560 | 5.76 |
ENSMUST00000021009.10
|
Myt1l
|
myelin transcription factor 1-like |
| chr4_-_151129020 | 5.67 |
ENSMUST00000103204.11
|
Per3
|
period circadian clock 3 |
| chr10_-_49659355 | 5.67 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr12_-_41536430 | 5.65 |
ENSMUST00000043884.6
|
Lrrn3
|
leucine rich repeat protein 3, neuronal |
| chr16_-_22258469 | 5.61 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr4_+_41941572 | 5.57 |
ENSMUST00000108028.9
ENSMUST00000153997.8 |
Gm20878
|
predicted gene, 20878 |
| chr17_-_90395771 | 5.53 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr10_-_125144678 | 5.51 |
ENSMUST00000105257.4
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr12_+_37291632 | 5.47 |
ENSMUST00000049874.14
|
Agmo
|
alkylglycerol monooxygenase |
| chr2_-_131170902 | 5.46 |
ENSMUST00000110194.8
|
Rnf24
|
ring finger protein 24 |
| chr3_-_123483772 | 5.44 |
ENSMUST00000172537.3
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr12_+_37292029 | 5.40 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr3_-_98660781 | 5.24 |
ENSMUST00000094050.11
ENSMUST00000090743.13 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr15_-_76191301 | 5.17 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr3_+_141171322 | 5.16 |
ENSMUST00000075282.10
|
Unc5c
|
unc-5 netrin receptor C |
| chr7_+_91711600 | 5.12 |
ENSMUST00000098308.4
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr5_-_130053120 | 5.12 |
ENSMUST00000161640.8
ENSMUST00000161884.2 ENSMUST00000161094.8 |
Asl
|
argininosuccinate lyase |
| chr14_+_4230658 | 4.94 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr1_+_55091922 | 4.89 |
ENSMUST00000087617.11
ENSMUST00000027125.7 |
Coq10b
|
coenzyme Q10B |
| chr10_-_6930376 | 4.89 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr13_+_16186410 | 4.86 |
ENSMUST00000042603.14
|
Inhba
|
inhibin beta-A |
| chr7_+_90125890 | 4.80 |
ENSMUST00000208919.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr4_-_110143777 | 4.80 |
ENSMUST00000138972.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr9_-_78254422 | 4.76 |
ENSMUST00000034902.12
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr10_+_23727325 | 4.74 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr6_+_103674695 | 4.70 |
ENSMUST00000205098.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr16_+_3702493 | 4.65 |
ENSMUST00000176233.2
|
Gm20695
|
predicted gene 20695 |
| chr9_-_78254443 | 4.60 |
ENSMUST00000129247.2
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr19_+_34560922 | 4.50 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr16_+_3702604 | 4.46 |
ENSMUST00000115860.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr18_+_37875135 | 4.45 |
ENSMUST00000003599.9
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr5_-_5564730 | 4.42 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr10_+_86599836 | 4.40 |
ENSMUST00000218802.2
|
Gm49358
|
predicted gene, 49358 |
| chr8_-_93806593 | 4.37 |
ENSMUST00000109582.3
|
Ces1b
|
carboxylesterase 1B |
| chrX_-_166907286 | 4.33 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr8_-_91860655 | 4.27 |
ENSMUST00000125257.3
|
Aktip
|
AKT interacting protein |
| chr18_+_37130860 | 4.26 |
ENSMUST00000115659.6
|
Pcdha9
|
protocadherin alpha 9 |
| chr2_-_131953359 | 4.24 |
ENSMUST00000128899.2
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr19_+_4560500 | 4.23 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr4_+_43383449 | 4.22 |
ENSMUST00000135216.2
ENSMUST00000152322.8 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr16_+_3702523 | 4.22 |
ENSMUST00000176625.8
ENSMUST00000186375.8 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr15_-_93234681 | 4.18 |
ENSMUST00000080299.7
|
Yaf2
|
YY1 associated factor 2 |
| chr7_+_27353331 | 4.17 |
ENSMUST00000008088.9
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr2_+_85715984 | 4.12 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr9_+_32305259 | 4.08 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr4_+_42438970 | 3.99 |
ENSMUST00000238328.2
|
Gm21586
|
predicted gene, 21586 |
| chr4_-_47474283 | 3.83 |
ENSMUST00000044148.3
|
Alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr5_-_145406533 | 3.81 |
ENSMUST00000031633.5
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr11_-_101676076 | 3.77 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr9_-_54554483 | 3.73 |
ENSMUST00000128163.8
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr18_+_37063237 | 3.72 |
ENSMUST00000193839.6
ENSMUST00000070797.7 |
Pcdha1
|
protocadherin alpha 1 |
| chr10_-_63926044 | 3.69 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_+_114697710 | 3.65 |
ENSMUST00000081752.13
|
Olfm3
|
olfactomedin 3 |
| chr7_-_103832599 | 3.61 |
ENSMUST00000216612.3
|
Olfr648
|
olfactory receptor 648 |
| chr8_+_46944000 | 3.59 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr18_+_37125424 | 3.58 |
ENSMUST00000194038.2
|
Pcdha8
|
protocadherin alpha 8 |
| chr19_-_59931432 | 3.58 |
ENSMUST00000170819.2
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I) |
| chr11_-_83959175 | 3.55 |
ENSMUST00000100705.11
|
Dusp14
|
dual specificity phosphatase 14 |
| chr12_+_37291728 | 3.54 |
ENSMUST00000160768.8
|
Agmo
|
alkylglycerol monooxygenase |
| chr5_-_5564873 | 3.49 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr15_+_101152078 | 3.49 |
ENSMUST00000228985.2
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr2_+_54326329 | 3.48 |
ENSMUST00000112636.8
ENSMUST00000112635.8 ENSMUST00000112634.8 |
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr2_-_60114684 | 3.48 |
ENSMUST00000028356.9
ENSMUST00000074606.11 |
Cd302
|
CD302 antigen |
| chr12_+_36431449 | 3.41 |
ENSMUST00000221452.2
ENSMUST00000062041.6 ENSMUST00000220519.2 ENSMUST00000221895.2 |
Crppa
|
CDP-L-ribitol pyrophosphorylase A |
| chr17_+_47604995 | 3.38 |
ENSMUST00000190020.4
|
Trerf1
|
transcriptional regulating factor 1 |
| chr2_-_130884602 | 3.36 |
ENSMUST00000028787.12
ENSMUST00000110239.8 ENSMUST00000110234.8 |
Gfra4
|
glial cell line derived neurotrophic factor family receptor alpha 4 |
| chr1_-_14380327 | 3.35 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr15_+_16728842 | 3.30 |
ENSMUST00000228307.2
|
Cdh9
|
cadherin 9 |
| chr8_-_91860576 | 3.29 |
ENSMUST00000120213.9
ENSMUST00000109609.9 |
Aktip
|
AKT interacting protein |
| chr12_+_87247297 | 3.24 |
ENSMUST00000182869.2
|
Samd15
|
sterile alpha motif domain containing 15 |
| chr15_-_37007623 | 3.19 |
ENSMUST00000078976.9
|
Zfp706
|
zinc finger protein 706 |
| chrX_-_74426221 | 3.10 |
ENSMUST00000147349.8
|
F8
|
coagulation factor VIII |
| chr8_-_93924426 | 3.09 |
ENSMUST00000034172.8
|
Ces1d
|
carboxylesterase 1D |
| chr11_-_115977755 | 3.08 |
ENSMUST00000074628.13
ENSMUST00000106444.4 |
Wbp2
|
WW domain binding protein 2 |
| chr10_+_80134917 | 3.08 |
ENSMUST00000154212.8
|
Apc2
|
APC regulator of WNT signaling pathway 2 |
| chr1_-_14380418 | 3.04 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr9_+_40180497 | 3.03 |
ENSMUST00000049941.12
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr9_+_78099229 | 3.02 |
ENSMUST00000034903.7
|
Gsta4
|
glutathione S-transferase, alpha 4 |
| chr5_+_67126304 | 3.00 |
ENSMUST00000132991.5
|
Limch1
|
LIM and calponin homology domains 1 |
| chr17_-_80885197 | 2.92 |
ENSMUST00000234602.2
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr2_-_20948230 | 2.92 |
ENSMUST00000140230.2
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr3_+_84859453 | 2.89 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr16_-_59138611 | 2.86 |
ENSMUST00000216261.2
|
Olfr204
|
olfactory receptor 204 |
| chr11_-_106107132 | 2.82 |
ENSMUST00000002043.10
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr1_-_38937190 | 2.82 |
ENSMUST00000193435.6
ENSMUST00000194361.6 |
Chst10
|
carbohydrate sulfotransferase 10 |
| chr10_+_20046748 | 2.80 |
ENSMUST00000215924.2
|
Map7
|
microtubule-associated protein 7 |
| chr15_+_7159038 | 2.77 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr9_+_40180569 | 2.75 |
ENSMUST00000176185.8
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr4_+_42293921 | 2.75 |
ENSMUST00000238203.2
|
Gm21953
|
predicted gene, 21953 |
| chr4_+_43493344 | 2.74 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr8_+_22550727 | 2.72 |
ENSMUST00000072572.13
ENSMUST00000110737.3 ENSMUST00000131624.2 |
Alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr2_+_118692435 | 2.71 |
ENSMUST00000028807.6
|
Ivd
|
isovaleryl coenzyme A dehydrogenase |
| chr5_+_117979899 | 2.71 |
ENSMUST00000142742.9
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr19_-_60849828 | 2.70 |
ENSMUST00000135808.8
|
Sfxn4
|
sideroflexin 4 |
| chr15_+_41694317 | 2.70 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr4_+_40473130 | 2.63 |
ENSMUST00000179526.2
|
Tmem215
|
transmembrane protein 215 |
| chr14_+_3576275 | 2.61 |
ENSMUST00000151926.8
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chrX_+_111510223 | 2.56 |
ENSMUST00000113409.8
|
Zfp711
|
zinc finger protein 711 |
| chr5_-_148931957 | 2.56 |
ENSMUST00000147473.6
|
Gm42791
|
predicted gene 42791 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.8 | 77.5 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 15.2 | 91.5 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 11.6 | 34.7 | GO:0097037 | heme export(GO:0097037) |
| 7.6 | 22.8 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 6.1 | 18.2 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 5.0 | 15.0 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 4.7 | 28.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 4.6 | 32.5 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 4.5 | 22.5 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 4.4 | 26.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 3.9 | 27.5 | GO:0048840 | otolith development(GO:0048840) |
| 3.7 | 11.0 | GO:0015881 | creatine transport(GO:0015881) |
| 3.6 | 10.7 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 3.4 | 20.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 3.0 | 23.9 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 2.9 | 8.6 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 2.2 | 6.6 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 2.2 | 6.6 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 2.1 | 19.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 2.1 | 8.5 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 2.0 | 5.9 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.9 | 26.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.9 | 62.7 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 1.8 | 7.2 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 1.8 | 19.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 1.7 | 5.1 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 1.7 | 8.5 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 1.6 | 16.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 1.6 | 6.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 1.4 | 34.0 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 1.4 | 4.2 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.4 | 9.9 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 1.1 | 9.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 1.1 | 14.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.0 | 34.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 1.0 | 7.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.0 | 3.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.0 | 4.9 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 1.0 | 8.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.9 | 5.5 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.9 | 23.8 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.8 | 4.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.8 | 9.9 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.8 | 11.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.7 | 2.7 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.7 | 7.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.7 | 2.0 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.6 | 9.0 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.6 | 1.9 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.6 | 5.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.6 | 4.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.6 | 2.9 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.6 | 2.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.6 | 8.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 3.8 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.5 | 1.6 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.5 | 1.6 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.5 | 2.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.5 | 5.8 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.5 | 1.6 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.5 | 6.8 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.5 | 3.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.5 | 1.5 | GO:1904268 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.5 | 4.6 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.5 | 6.0 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.5 | 9.9 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.5 | 1.9 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.5 | 1.5 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.5 | 4.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.5 | 13.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.4 | 3.5 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.4 | 1.6 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.4 | 11.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.4 | 13.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.4 | 3.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.4 | 6.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.4 | 6.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.4 | 7.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.4 | 11.5 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.4 | 2.5 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 2.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.3 | 2.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 3.6 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.3 | 1.6 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.3 | 1.8 | GO:1903758 | regulation of postsynaptic density protein 95 clustering(GO:1902897) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.3 | 2.3 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.3 | 3.7 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.3 | 2.7 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.3 | 1.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 1.9 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 9.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 1.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 23.2 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.2 | 1.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 4.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 1.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 2.8 | GO:0006983 | ER overload response(GO:0006983) |
| 0.2 | 2.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 1.9 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 | 5.7 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 7.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 1.8 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 2.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 1.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 2.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 2.7 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 8.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 0.8 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 0.6 | GO:1902267 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 3.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 2.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 9.8 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 7.6 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 7.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 3.1 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.1 | 7.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 4.7 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 18.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 5.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 3.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 2.8 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 1.8 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 4.7 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 9.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.1 | 0.6 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 1.8 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 1.3 | GO:0046174 | inositol metabolic process(GO:0006020) polyol catabolic process(GO:0046174) |
| 0.1 | 3.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 2.4 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.8 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 3.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 5.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.4 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.6 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 5.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 2.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 1.5 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 9.9 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 | 3.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 | 0.9 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 1.2 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 1.7 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 1.1 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 1.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 3.6 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) |
| 0.0 | 4.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 5.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 3.4 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 6.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 8.6 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 5.6 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 0.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 4.5 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 3.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 2.6 | GO:0010771 | negative regulation of cell morphogenesis involved in differentiation(GO:0010771) |
| 0.0 | 3.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 2.3 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.6 | 34.7 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 6.1 | 91.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.6 | 4.9 | GO:0043512 | inhibin A complex(GO:0043512) |
| 1.3 | 12.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 1.1 | 29.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.1 | 7.6 | GO:0070695 | FHF complex(GO:0070695) |
| 1.0 | 27.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.0 | 13.4 | GO:0005883 | neurofilament(GO:0005883) |
| 1.0 | 8.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.9 | 8.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.8 | 6.2 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.7 | 2.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.7 | 5.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.5 | 50.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.5 | 77.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.5 | 7.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.4 | 5.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.4 | 1.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.3 | 9.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 8.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 5.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 37.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.3 | 13.6 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.2 | 29.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 1.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 5.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 2.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 3.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 20.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 60.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 1.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 5.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 2.7 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 41.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 40.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 4.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 3.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 36.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 5.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 7.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 31.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 2.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 19.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 6.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 3.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 5.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 3.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 6.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 3.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 9.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 2.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 16.5 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 4.3 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 1.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.7 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 21.0 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 4.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 9.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 37.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 9.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 6.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 5.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 13.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.4 | 77.5 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 11.3 | 34.0 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 7.5 | 22.5 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 7.1 | 28.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 6.5 | 6.5 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 5.4 | 32.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 5.1 | 20.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 4.8 | 14.4 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 4.6 | 37.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 4.5 | 18.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 3.9 | 27.5 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 3.7 | 11.0 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 3.5 | 91.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 2.9 | 23.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 2.8 | 11.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 2.4 | 19.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 2.4 | 7.1 | GO:0016232 | HNK-1 sulfotransferase activity(GO:0016232) |
| 2.2 | 6.6 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 2.2 | 13.2 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 2.1 | 10.7 | GO:1903135 | cupric ion binding(GO:1903135) |
| 2.0 | 5.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.9 | 13.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.4 | 4.2 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.3 | 5.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.3 | 3.8 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.2 | 11.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.2 | 17.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 1.2 | 18.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 1.1 | 9.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.1 | 32.6 | GO:0031402 | sodium ion binding(GO:0031402) |
| 1.1 | 4.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.0 | 7.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.0 | 3.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.0 | 36.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.9 | 5.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.9 | 11.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.8 | 15.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.8 | 9.9 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.8 | 2.5 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.8 | 5.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.8 | 5.4 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.8 | 3.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.7 | 9.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.7 | 11.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.7 | 5.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.7 | 3.6 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.7 | 4.9 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.7 | 4.9 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.7 | 2.7 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.7 | 6.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.7 | 3.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.7 | 8.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.6 | 2.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.6 | 4.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 8.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.6 | 5.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.5 | 2.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.5 | 4.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.5 | 2.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.5 | 3.8 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.5 | 2.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.4 | 3.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 2.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.4 | 1.6 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 2.4 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.4 | 9.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 8.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 1.6 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.4 | 6.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 5.8 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.3 | 9.3 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 7.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 9.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 31.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.3 | 13.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.3 | 2.0 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.3 | 0.8 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.3 | 1.9 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.3 | 1.3 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.3 | 3.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.3 | 29.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 1.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 9.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 3.7 | GO:0102391 | very long-chain fatty acid-CoA ligase activity(GO:0031957) decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 37.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 2.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 1.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 6.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 2.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 0.6 | GO:0042979 | ornithine decarboxylase inhibitor activity(GO:0008073) ornithine decarboxylase regulator activity(GO:0042979) |
| 0.2 | 1.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.5 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.8 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 1.7 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.8 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 7.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 2.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 4.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 6.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.6 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 13.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 0.9 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 13.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 9.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.9 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 2.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 4.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 2.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 8.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 3.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 3.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 3.3 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 10.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 12.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 9.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 10.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 4.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 1.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 8.6 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 1.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 3.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 74.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.6 | 15.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 23.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.3 | 22.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 21.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 12.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 10.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 10.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 13.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 19.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 4.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 4.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 14.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 6.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 7.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 19.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 59.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 2.3 | 77.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 1.3 | 48.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 1.2 | 27.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 1.1 | 9.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.0 | 21.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.0 | 129.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.6 | 25.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.5 | 19.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.4 | 13.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 33.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.4 | 5.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 21.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 15.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.3 | 14.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 9.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 14.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 8.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 7.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 2.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 8.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 6.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 15.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 10.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 5.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 7.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.3 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 3.6 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 2.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 4.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 3.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 3.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.6 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 3.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 2.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 5.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 3.3 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 3.1 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |