Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Nfix
Z-value: 1.58
Transcription factors associated with Nfix
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfix
|
ENSMUSG00000001911.17 | Nfix |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfix | mm39_v1_chr8_-_85500998_85501108 | 0.72 | 1.6e-12 | Click! |
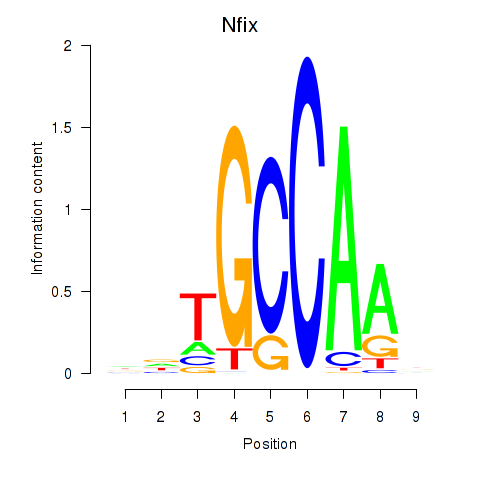
Activity profile of Nfix motif
Sorted Z-values of Nfix motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfix
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_63006176 | 19.36 |
ENSMUST00000021907.9
|
Fbp2
|
fructose bisphosphatase 2 |
| chr7_+_130633776 | 12.30 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr7_+_130179063 | 12.21 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr11_-_100012384 | 10.80 |
ENSMUST00000007275.3
|
Krt13
|
keratin 13 |
| chr3_-_69767208 | 10.29 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr5_+_115604321 | 9.54 |
ENSMUST00000145785.8
ENSMUST00000031495.11 ENSMUST00000112071.8 ENSMUST00000125568.2 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr17_-_82045800 | 9.49 |
ENSMUST00000235015.2
ENSMUST00000163123.3 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr5_-_87054796 | 9.46 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr5_-_24829395 | 8.73 |
ENSMUST00000195943.2
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr12_-_76842263 | 8.50 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr3_-_144738526 | 8.38 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr15_-_101833160 | 7.90 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr11_+_52265090 | 7.61 |
ENSMUST00000020673.3
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr14_+_70694887 | 7.45 |
ENSMUST00000003561.10
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein |
| chr13_+_49761506 | 7.35 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr12_+_36042899 | 7.27 |
ENSMUST00000020898.12
|
Agr2
|
anterior gradient 2 |
| chr11_+_3438274 | 7.10 |
ENSMUST00000064265.13
|
Pla2g3
|
phospholipase A2, group III |
| chr3_+_87855973 | 6.97 |
ENSMUST00000005019.6
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr2_-_163239865 | 6.65 |
ENSMUST00000017961.11
ENSMUST00000109425.3 |
Jph2
|
junctophilin 2 |
| chr4_-_155095441 | 6.51 |
ENSMUST00000105631.9
ENSMUST00000139976.9 ENSMUST00000145662.9 |
Plch2
|
phospholipase C, eta 2 |
| chr16_+_78727829 | 6.40 |
ENSMUST00000114216.2
ENSMUST00000069148.13 ENSMUST00000023568.14 |
Chodl
|
chondrolectin |
| chr11_-_83959175 | 6.39 |
ENSMUST00000100705.11
|
Dusp14
|
dual specificity phosphatase 14 |
| chr11_+_98727611 | 6.23 |
ENSMUST00000107479.3
|
Rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr16_+_20513658 | 5.98 |
ENSMUST00000056518.13
|
Fam131a
|
family with sequence similarity 131, member A |
| chr19_+_24651362 | 5.95 |
ENSMUST00000057243.6
|
Tmem252
|
transmembrane protein 252 |
| chr7_-_126651847 | 5.84 |
ENSMUST00000205424.2
|
Zg16
|
zymogen granule protein 16 |
| chr8_+_95498822 | 5.70 |
ENSMUST00000211956.2
ENSMUST00000211947.2 |
Cx3cl1
|
chemokine (C-X3-C motif) ligand 1 |
| chr8_-_95405234 | 5.52 |
ENSMUST00000213043.2
|
Pllp
|
plasma membrane proteolipid |
| chr1_-_128256048 | 5.50 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr6_+_141470105 | 5.47 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr11_-_83959380 | 5.43 |
ENSMUST00000164891.8
|
Dusp14
|
dual specificity phosphatase 14 |
| chr5_+_149335214 | 5.43 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr11_+_54205722 | 5.27 |
ENSMUST00000072178.11
ENSMUST00000101211.9 ENSMUST00000101213.9 ENSMUST00000064690.10 ENSMUST00000108899.8 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr15_-_78090591 | 5.27 |
ENSMUST00000120592.2
|
Pvalb
|
parvalbumin |
| chrX_+_10351360 | 5.18 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr10_-_89093441 | 5.17 |
ENSMUST00000182341.8
ENSMUST00000182613.8 |
Ano4
|
anoctamin 4 |
| chr7_+_26958150 | 4.97 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chr10_+_57660948 | 4.95 |
ENSMUST00000020024.12
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr12_-_72283465 | 4.85 |
ENSMUST00000021497.16
ENSMUST00000137990.2 |
Rtn1
|
reticulon 1 |
| chr7_+_126396779 | 4.81 |
ENSMUST00000205324.2
|
Tlcd3b
|
TLC domain containing 3B |
| chr4_-_141351110 | 4.81 |
ENSMUST00000038661.8
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr1_+_87522267 | 4.63 |
ENSMUST00000165109.2
ENSMUST00000070898.6 |
Neu2
|
neuraminidase 2 |
| chr9_+_22012357 | 4.63 |
ENSMUST00000216872.2
|
Cnn1
|
calponin 1 |
| chr1_+_164624200 | 4.58 |
ENSMUST00000027861.6
|
Dpt
|
dermatopontin |
| chr2_+_72128239 | 4.57 |
ENSMUST00000144111.2
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr18_+_44096300 | 4.43 |
ENSMUST00000069245.8
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr14_-_118474404 | 4.39 |
ENSMUST00000170662.2
|
Sox21
|
SRY (sex determining region Y)-box 21 |
| chr1_+_171077984 | 4.34 |
ENSMUST00000111315.9
ENSMUST00000219033.2 |
Adamts4
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 4 |
| chr16_+_41353212 | 4.27 |
ENSMUST00000078873.11
|
Lsamp
|
limbic system-associated membrane protein |
| chr11_+_96822213 | 4.26 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr11_+_68986043 | 4.25 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr15_-_37458768 | 4.24 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr15_-_101621332 | 4.23 |
ENSMUST00000023709.7
|
Krt5
|
keratin 5 |
| chr12_-_72455708 | 4.22 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr3_-_127202693 | 4.09 |
ENSMUST00000182078.9
|
Ank2
|
ankyrin 2, brain |
| chr11_+_63019799 | 4.09 |
ENSMUST00000108702.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr13_+_43019718 | 4.07 |
ENSMUST00000131942.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr10_+_116013256 | 4.06 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr11_+_69047815 | 4.06 |
ENSMUST00000036424.3
|
Alox12b
|
arachidonate 12-lipoxygenase, 12R type |
| chr3_+_64884839 | 4.06 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr8_+_31581635 | 4.06 |
ENSMUST00000161713.2
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr11_+_118878999 | 4.05 |
ENSMUST00000092373.13
ENSMUST00000106273.3 |
Enpp7
|
ectonucleotide pyrophosphatase/phosphodiesterase 7 |
| chr15_-_64794139 | 3.97 |
ENSMUST00000023007.7
ENSMUST00000228014.2 |
Adcy8
|
adenylate cyclase 8 |
| chr14_-_55150547 | 3.89 |
ENSMUST00000228495.3
ENSMUST00000228119.3 ENSMUST00000050772.10 ENSMUST00000231305.2 |
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr8_-_120362291 | 3.88 |
ENSMUST00000061828.10
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr11_+_67090878 | 3.88 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr6_+_135339929 | 3.83 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr2_+_35512023 | 3.83 |
ENSMUST00000091010.12
|
Dab2ip
|
disabled 2 interacting protein |
| chr1_-_172047282 | 3.79 |
ENSMUST00000170700.2
ENSMUST00000003554.11 |
Casq1
|
calsequestrin 1 |
| chr10_+_57661010 | 3.72 |
ENSMUST00000165013.2
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr3_+_99203818 | 3.71 |
ENSMUST00000150756.3
|
Tbx15
|
T-box 15 |
| chr18_-_20247824 | 3.70 |
ENSMUST00000038710.6
|
Dsc1
|
desmocollin 1 |
| chr6_+_78347636 | 3.70 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr18_+_37840092 | 3.65 |
ENSMUST00000195823.2
|
Pcdhga6
|
protocadherin gamma subfamily A, 6 |
| chr9_-_60418286 | 3.65 |
ENSMUST00000098660.10
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr7_+_81984940 | 3.59 |
ENSMUST00000173287.8
|
Adamtsl3
|
ADAMTS-like 3 |
| chr6_-_115228800 | 3.53 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr12_+_84408803 | 3.52 |
ENSMUST00000110278.8
ENSMUST00000145522.2 |
Coq6
|
coenzyme Q6 monooxygenase |
| chr12_-_70394074 | 3.44 |
ENSMUST00000223160.2
ENSMUST00000222316.2 ENSMUST00000167755.3 ENSMUST00000110520.10 ENSMUST00000110522.10 ENSMUST00000221041.2 ENSMUST00000222603.3 |
Trim9
|
tripartite motif-containing 9 |
| chr18_-_23171713 | 3.42 |
ENSMUST00000081423.13
|
Nol4
|
nucleolar protein 4 |
| chr7_-_142223662 | 3.38 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr1_+_66360865 | 3.37 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr5_-_77262968 | 3.34 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr2_-_113883285 | 3.33 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr13_-_113800172 | 3.32 |
ENSMUST00000054650.5
|
Hspb3
|
heat shock protein 3 |
| chr6_-_24956296 | 3.31 |
ENSMUST00000127247.4
|
Tmem229a
|
transmembrane protein 229A |
| chr13_-_32960379 | 3.31 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr2_-_25471703 | 3.30 |
ENSMUST00000114217.3
ENSMUST00000191602.2 |
Ajm1
|
apical junction component 1 |
| chr6_-_124790029 | 3.28 |
ENSMUST00000149610.3
|
Tpi1
|
triosephosphate isomerase 1 |
| chr5_-_137919873 | 3.24 |
ENSMUST00000031741.8
|
Cyp3a13
|
cytochrome P450, family 3, subfamily a, polypeptide 13 |
| chr11_-_114686712 | 3.23 |
ENSMUST00000000206.4
|
Btbd17
|
BTB (POZ) domain containing 17 |
| chr10_+_116013122 | 3.20 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr13_+_93440265 | 3.19 |
ENSMUST00000109494.8
|
Homer1
|
homer scaffolding protein 1 |
| chr6_+_78347844 | 3.17 |
ENSMUST00000096904.6
ENSMUST00000203266.2 |
Reg3b
|
regenerating islet-derived 3 beta |
| chr11_+_110914678 | 3.17 |
ENSMUST00000150902.8
ENSMUST00000178798.2 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr9_+_21095399 | 3.16 |
ENSMUST00000115458.9
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr11_+_16207705 | 3.14 |
ENSMUST00000109645.9
ENSMUST00000109647.3 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chrX_-_58613428 | 3.13 |
ENSMUST00000119833.8
ENSMUST00000131319.8 |
Fgf13
|
fibroblast growth factor 13 |
| chr2_+_51928017 | 3.11 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr12_-_84408576 | 3.07 |
ENSMUST00000021659.2
ENSMUST00000065536.9 |
Fam161b
|
family with sequence similarity 161, member B |
| chr1_+_152275575 | 3.00 |
ENSMUST00000044311.9
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr3_+_62245765 | 2.91 |
ENSMUST00000079300.13
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr12_-_45120895 | 2.90 |
ENSMUST00000120531.8
ENSMUST00000143376.8 |
Stxbp6
|
syntaxin binding protein 6 (amisyn) |
| chr7_+_141276575 | 2.89 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr1_+_133109059 | 2.88 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr11_-_102787950 | 2.85 |
ENSMUST00000067444.10
|
Gfap
|
glial fibrillary acidic protein |
| chr2_+_22785534 | 2.82 |
ENSMUST00000053729.14
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr11_-_102787972 | 2.81 |
ENSMUST00000077902.5
|
Gfap
|
glial fibrillary acidic protein |
| chr7_+_27353331 | 2.79 |
ENSMUST00000008088.9
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr3_-_127202635 | 2.79 |
ENSMUST00000182959.8
|
Ank2
|
ankyrin 2, brain |
| chr13_+_120151982 | 2.78 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr18_-_20247928 | 2.76 |
ENSMUST00000226115.2
|
Dsc1
|
desmocollin 1 |
| chr18_-_20247666 | 2.75 |
ENSMUST00000224432.2
|
Dsc1
|
desmocollin 1 |
| chr6_+_135339543 | 2.75 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr19_-_40576817 | 2.71 |
ENSMUST00000175932.2
ENSMUST00000176955.8 ENSMUST00000149476.3 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr12_+_84408742 | 2.71 |
ENSMUST00000021661.13
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr1_-_86039692 | 2.70 |
ENSMUST00000027431.7
|
Htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B |
| chr1_+_159351337 | 2.66 |
ENSMUST00000192069.6
|
Tnr
|
tenascin R |
| chr6_-_102441628 | 2.66 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr13_+_42862957 | 2.65 |
ENSMUST00000066928.12
ENSMUST00000148891.8 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr15_+_10952418 | 2.63 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr13_+_120151915 | 2.59 |
ENSMUST00000225543.2
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr3_-_89186940 | 2.58 |
ENSMUST00000118860.2
ENSMUST00000029566.9 |
Efna1
|
ephrin A1 |
| chr11_+_115802828 | 2.57 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr8_-_65186565 | 2.56 |
ENSMUST00000141021.2
|
Msmo1
|
methylsterol monoxygenase 1 |
| chr2_+_20742115 | 2.55 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr11_+_98274637 | 2.55 |
ENSMUST00000008021.3
|
Tcap
|
titin-cap |
| chr2_-_24985161 | 2.54 |
ENSMUST00000044018.8
|
Noxa1
|
NADPH oxidase activator 1 |
| chr3_+_107009896 | 2.54 |
ENSMUST00000196403.2
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr1_+_169757498 | 2.53 |
ENSMUST00000175731.3
|
Ccdc190
|
coiled-coil domain containing 190 |
| chr7_+_44499374 | 2.52 |
ENSMUST00000141311.8
ENSMUST00000107880.9 ENSMUST00000208384.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr3_+_123240562 | 2.50 |
ENSMUST00000029603.10
|
Prss12
|
protease, serine 12 neurotrypsin (motopsin) |
| chr13_-_23806530 | 2.49 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr3_-_127202663 | 2.48 |
ENSMUST00000182008.8
ENSMUST00000182547.8 |
Ank2
|
ankyrin 2, brain |
| chr2_+_22785593 | 2.47 |
ENSMUST00000152170.8
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr5_+_92535532 | 2.43 |
ENSMUST00000145072.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chrX_+_113384008 | 2.41 |
ENSMUST00000113371.8
ENSMUST00000040504.12 |
Klhl4
|
kelch-like 4 |
| chr5_+_148202011 | 2.40 |
ENSMUST00000110515.9
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr9_-_106324642 | 2.39 |
ENSMUST00000185334.7
ENSMUST00000187001.2 ENSMUST00000171678.9 ENSMUST00000190798.7 ENSMUST00000048685.13 ENSMUST00000171925.8 |
Abhd14a
|
abhydrolase domain containing 14A |
| chrX_-_166638057 | 2.39 |
ENSMUST00000238211.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chrX_-_20816841 | 2.37 |
ENSMUST00000009550.14
|
Elk1
|
ELK1, member of ETS oncogene family |
| chr3_+_76500857 | 2.34 |
ENSMUST00000162471.2
|
Fstl5
|
follistatin-like 5 |
| chr9_+_89791943 | 2.33 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr2_-_24985137 | 2.30 |
ENSMUST00000114373.8
|
Noxa1
|
NADPH oxidase activator 1 |
| chr18_-_47466378 | 2.30 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr18_-_65527078 | 2.28 |
ENSMUST00000035548.16
|
Alpk2
|
alpha-kinase 2 |
| chr5_-_31250817 | 2.27 |
ENSMUST00000031037.14
|
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr19_+_8828132 | 2.25 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chrX_+_105230706 | 2.21 |
ENSMUST00000081593.13
|
Pgk1
|
phosphoglycerate kinase 1 |
| chr7_+_19699291 | 2.20 |
ENSMUST00000094753.6
|
Ceacam20
|
carcinoembryonic antigen-related cell adhesion molecule 20 |
| chr10_+_21869776 | 2.18 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr11_+_78181667 | 2.18 |
ENSMUST00000046361.5
ENSMUST00000238934.2 |
Rskr
|
ribosomal protein S6 kinase related |
| chr17_-_49871291 | 2.16 |
ENSMUST00000224595.2
ENSMUST00000057610.8 |
Daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr3_-_108322868 | 2.16 |
ENSMUST00000090558.10
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chrX_+_103526380 | 2.15 |
ENSMUST00000087867.6
|
Uprt
|
uracil phosphoribosyltransferase |
| chr19_-_40576897 | 2.13 |
ENSMUST00000025979.13
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr5_+_92535705 | 2.12 |
ENSMUST00000138687.2
ENSMUST00000124509.2 |
Art3
|
ADP-ribosyltransferase 3 |
| chr12_+_119354110 | 2.11 |
ENSMUST00000222058.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr8_+_59364789 | 2.11 |
ENSMUST00000062978.7
|
BC030500
|
cDNA sequence BC030500 |
| chr5_-_84565218 | 2.09 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr13_+_83720457 | 2.09 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr18_+_4994600 | 2.06 |
ENSMUST00000140448.8
|
Svil
|
supervillin |
| chr18_+_38088179 | 2.04 |
ENSMUST00000176902.8
ENSMUST00000176104.8 |
Rell2
|
RELT-like 2 |
| chr6_-_33037107 | 2.04 |
ENSMUST00000115091.2
ENSMUST00000127666.8 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr19_-_43512929 | 2.03 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chrX_-_105647282 | 2.02 |
ENSMUST00000113480.2
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr7_-_19621833 | 1.97 |
ENSMUST00000052605.8
|
Ceacam19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr6_-_136852792 | 1.96 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chrX_+_149829131 | 1.93 |
ENSMUST00000112685.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr9_+_121196034 | 1.93 |
ENSMUST00000210636.2
ENSMUST00000045903.8 |
Trak1
|
trafficking protein, kinesin binding 1 |
| chr11_+_50917831 | 1.93 |
ENSMUST00000072152.2
|
Olfr54
|
olfactory receptor 54 |
| chr2_+_133394079 | 1.91 |
ENSMUST00000028836.7
|
Bmp2
|
bone morphogenetic protein 2 |
| chr11_+_67128843 | 1.87 |
ENSMUST00000018632.11
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr19_-_40576782 | 1.85 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr8_-_120362085 | 1.83 |
ENSMUST00000164382.2
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr3_-_127202586 | 1.82 |
ENSMUST00000183095.3
ENSMUST00000182610.8 |
Ank2
|
ankyrin 2, brain |
| chr3_+_128993568 | 1.82 |
ENSMUST00000029657.16
ENSMUST00000106382.11 |
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr2_+_3115250 | 1.81 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr7_+_120442048 | 1.79 |
ENSMUST00000047875.16
|
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr18_-_43032359 | 1.78 |
ENSMUST00000117687.8
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr7_+_44498640 | 1.73 |
ENSMUST00000054343.15
ENSMUST00000142880.3 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr13_+_83720484 | 1.72 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr19_-_40600619 | 1.72 |
ENSMUST00000132452.2
ENSMUST00000135795.8 ENSMUST00000025981.15 |
Tctn3
|
tectonic family member 3 |
| chr11_+_67689094 | 1.71 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr14_-_36628263 | 1.70 |
ENSMUST00000183007.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_+_74583504 | 1.66 |
ENSMUST00000027362.14
|
Plcd4
|
phospholipase C, delta 4 |
| chr9_+_43978369 | 1.66 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr12_-_91712783 | 1.62 |
ENSMUST00000166967.2
|
Ston2
|
stonin 2 |
| chr19_+_56276343 | 1.62 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr4_+_102287244 | 1.60 |
ENSMUST00000172616.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr4_-_129155185 | 1.57 |
ENSMUST00000145261.8
|
C77080
|
expressed sequence C77080 |
| chr18_+_69633741 | 1.56 |
ENSMUST00000207214.2
ENSMUST00000201094.4 ENSMUST00000200703.4 ENSMUST00000202765.4 |
Tcf4
|
transcription factor 4 |
| chr17_-_36012932 | 1.55 |
ENSMUST00000166980.9
ENSMUST00000145900.8 |
Ddr1
|
discoidin domain receptor family, member 1 |
| chr2_-_5680801 | 1.54 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr5_-_123887434 | 1.53 |
ENSMUST00000182955.8
ENSMUST00000182489.8 ENSMUST00000183147.9 ENSMUST00000050827.14 ENSMUST00000057795.12 ENSMUST00000111515.8 ENSMUST00000182309.8 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr16_+_20492014 | 1.50 |
ENSMUST00000154950.8
ENSMUST00000115461.8 ENSMUST00000136713.5 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr8_-_68658694 | 1.47 |
ENSMUST00000212960.2
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr3_+_89987749 | 1.46 |
ENSMUST00000127955.2
|
Tpm3
|
tropomyosin 3, gamma |
| chr13_+_25127127 | 1.45 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr1_+_90131702 | 1.45 |
ENSMUST00000065587.5
ENSMUST00000159654.2 |
Ackr3
|
atypical chemokine receptor 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 17.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 3.2 | 9.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 2.4 | 7.3 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 2.2 | 6.7 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 2.1 | 8.5 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 1.4 | 7.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 1.3 | 10.3 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 1.2 | 8.7 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.2 | 11.2 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.2 | 7.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.1 | 3.4 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 1.1 | 5.7 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.1 | 6.7 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 1.1 | 3.3 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.1 | 5.3 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 1.1 | 9.5 | GO:0098735 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.9 | 3.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.9 | 5.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.9 | 5.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.8 | 4.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.8 | 4.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.8 | 3.8 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.7 | 4.4 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.7 | 5.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.7 | 4.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.7 | 2.0 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.7 | 2.7 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.6 | 3.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.6 | 3.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.6 | 7.5 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.6 | 2.9 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.6 | 4.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.6 | 5.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.5 | 5.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.5 | 12.2 | GO:0030953 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.5 | 4.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.5 | 11.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.5 | 1.9 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.5 | 6.2 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.5 | 12.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.4 | 2.7 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.4 | 2.6 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) notochord formation(GO:0014028) |
| 0.4 | 2.5 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.4 | 4.6 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.4 | 7.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.4 | 7.6 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.4 | 10.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 1.9 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.4 | 5.0 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.4 | 2.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.3 | 2.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.3 | 8.7 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.3 | 2.6 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.3 | 2.3 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.3 | 3.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 6.4 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.3 | 3.3 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.3 | 1.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 2.1 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.3 | 2.4 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.3 | 3.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 0.8 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 11.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 0.8 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.2 | 4.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 0.9 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.2 | 2.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 2.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 3.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 4.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.2 | 5.4 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.2 | 0.8 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.2 | 4.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.7 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 1.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 2.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 2.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.7 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.1 | 3.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.9 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.1 | 2.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 5.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 2.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 5.1 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 8.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 4.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 1.3 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.3 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.9 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 2.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 1.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 4.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 4.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 1.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 | 1.2 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.1 | 0.6 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 2.1 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.1 | 0.2 | GO:0046436 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.1 | GO:2000729 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.3 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.1 | 0.3 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.1 | 2.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 9.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 5.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 2.4 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.5 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 3.7 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 5.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 5.6 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 1.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.0 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 2.3 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) negative regulation of centriole replication(GO:0046600) |
| 0.0 | 6.6 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0021763 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 2.2 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 1.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.0 | 1.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 3.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.8 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 3.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 2.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 2.0 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 1.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 2.2 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 4.8 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.5 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.3 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 1.7 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 2.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.2 | 26.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 1.1 | 10.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 1.1 | 3.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 1.0 | 3.8 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 1.0 | 3.8 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.9 | 4.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.7 | 6.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.7 | 5.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.7 | 2.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.6 | 1.7 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.4 | 6.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.4 | 3.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.4 | 4.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.4 | 3.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 15.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 8.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 1.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 7.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.3 | 9.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.3 | 16.4 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 9.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 4.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 3.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 3.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 8.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 5.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 5.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 0.9 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 2.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 0.8 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 2.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 2.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 10.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 2.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 2.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 2.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.9 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 13.2 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 7.1 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 5.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 4.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 5.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 17.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 6.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 2.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 9.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 69.5 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 1.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 3.2 | 9.5 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 2.2 | 6.7 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 2.1 | 10.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.8 | 5.4 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.3 | 5.3 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 1.1 | 2.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 1.1 | 9.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 1.0 | 15.5 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 1.0 | 2.9 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.9 | 4.6 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.9 | 5.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.8 | 7.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.7 | 11.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.7 | 5.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.7 | 2.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.7 | 4.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.6 | 4.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.5 | 2.0 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.5 | 2.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 3.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.5 | 10.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 13.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.4 | 4.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.4 | 2.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 7.0 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 4.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 5.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 2.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.4 | 9.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 6.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 3.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 4.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 1.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.3 | 8.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 0.9 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.3 | 1.5 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.3 | 7.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 2.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 8.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 5.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.3 | 8.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 3.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.2 | 0.9 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 0.9 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 8.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.9 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.2 | 2.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 1.5 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.2 | 2.7 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 8.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 6.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 0.5 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.2 | 6.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 4.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 1.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 3.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 2.7 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 5.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 5.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 11.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 4.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 3.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 2.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 3.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 4.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 4.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 2.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 8.0 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 11.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 3.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 3.4 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 3.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 10.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 3.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 3.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.6 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 13.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 1.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 7.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 14.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 4.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 15.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 7.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 7.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 7.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 4.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 5.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 9.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 18.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 4.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 6.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 6.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.7 | 16.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.5 | 6.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 26.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 9.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 5.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 6.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 4.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 2.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 9.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 4.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 9.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 11.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 12.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 5.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 4.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 7.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 2.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 2.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.4 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.8 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 3.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |