Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
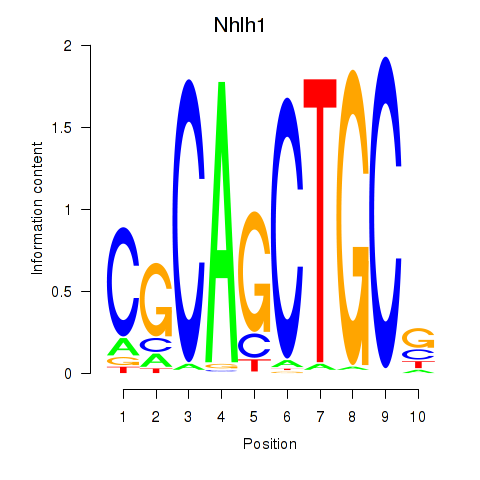
Results for Nhlh1
Z-value: 1.75
Transcription factors associated with Nhlh1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nhlh1
|
ENSMUSG00000051251.4 | Nhlh1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nhlh1 | mm39_v1_chr1_-_171885140_171885169 | 0.65 | 8.2e-10 | Click! |
Activity profile of Nhlh1 motif
Sorted Z-values of Nhlh1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nhlh1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_74435587 | 12.46 |
ENSMUST00000185682.7
ENSMUST00000170845.8 ENSMUST00000187599.2 |
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr13_+_92562404 | 12.41 |
ENSMUST00000061594.13
|
Ankrd34b
|
ankyrin repeat domain 34B |
| chr19_+_47003111 | 11.95 |
ENSMUST00000037636.4
|
Ina
|
internexin neuronal intermediate filament protein, alpha |
| chr17_-_26420300 | 11.64 |
ENSMUST00000025019.9
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr3_+_13536696 | 11.28 |
ENSMUST00000191806.3
ENSMUST00000193117.3 |
Ralyl
|
RALY RNA binding protein-like |
| chr7_+_121888520 | 10.47 |
ENSMUST00000064989.12
ENSMUST00000064921.5 |
Prkcb
|
protein kinase C, beta |
| chr1_+_174329361 | 10.42 |
ENSMUST00000030039.13
|
Fmn2
|
formin 2 |
| chr13_-_10410857 | 10.38 |
ENSMUST00000187510.7
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chr2_+_70392351 | 9.99 |
ENSMUST00000094934.11
|
Gad1
|
glutamate decarboxylase 1 |
| chr17_-_26420332 | 9.82 |
ENSMUST00000121959.3
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr4_-_56990306 | 9.55 |
ENSMUST00000053681.6
|
Frrs1l
|
ferric-chelate reductase 1 like |
| chr17_+_81251997 | 9.29 |
ENSMUST00000025092.5
|
Tmem178
|
transmembrane protein 178 |
| chr2_-_180776920 | 9.16 |
ENSMUST00000197015.5
ENSMUST00000103050.10 ENSMUST00000081528.13 ENSMUST00000049792.15 ENSMUST00000103048.10 ENSMUST00000103047.10 ENSMUST00000149964.9 |
Kcnq2
|
potassium voltage-gated channel, subfamily Q, member 2 |
| chr9_+_107812873 | 9.05 |
ENSMUST00000035700.14
|
Camkv
|
CaM kinase-like vesicle-associated |
| chr2_+_158217558 | 9.05 |
ENSMUST00000109488.8
|
Snhg11
|
small nucleolar RNA host gene 11 |
| chr16_-_45544960 | 8.62 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr15_-_98705791 | 8.50 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr14_-_29443792 | 8.47 |
ENSMUST00000022567.9
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr16_+_91066602 | 8.45 |
ENSMUST00000056882.7
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr3_-_80820835 | 8.44 |
ENSMUST00000107743.8
ENSMUST00000029654.15 |
Glrb
|
glycine receptor, beta subunit |
| chr7_+_49624978 | 8.26 |
ENSMUST00000107603.2
|
Nell1
|
NEL-like 1 |
| chr7_-_57159119 | 8.23 |
ENSMUST00000206382.2
|
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr9_-_75518585 | 8.19 |
ENSMUST00000098552.10
ENSMUST00000064433.11 |
Tmod2
|
tropomodulin 2 |
| chr8_+_31581635 | 8.18 |
ENSMUST00000161713.2
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr7_-_30826376 | 8.18 |
ENSMUST00000098548.8
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr9_-_121323906 | 8.12 |
ENSMUST00000215228.2
ENSMUST00000213106.2 |
Cck
|
cholecystokinin |
| chr2_+_49509288 | 8.11 |
ENSMUST00000028102.14
|
Kif5c
|
kinesin family member 5C |
| chr2_-_25209107 | 8.09 |
ENSMUST00000114318.10
ENSMUST00000114310.10 ENSMUST00000114308.10 ENSMUST00000114317.10 ENSMUST00000028335.13 ENSMUST00000114314.10 ENSMUST00000114307.8 |
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr10_-_79473675 | 8.09 |
ENSMUST00000020564.7
|
Shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr9_+_121589044 | 8.06 |
ENSMUST00000093772.4
|
Zfp651
|
zinc finger protein 651 |
| chr8_-_112120442 | 8.03 |
ENSMUST00000038475.9
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr15_-_75438457 | 7.97 |
ENSMUST00000163116.8
ENSMUST00000023241.12 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr5_-_139115914 | 7.96 |
ENSMUST00000129851.8
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chr11_-_119438569 | 7.95 |
ENSMUST00000026670.5
|
Nptx1
|
neuronal pentraxin 1 |
| chr7_-_84021655 | 7.80 |
ENSMUST00000208392.2
ENSMUST00000208232.2 ENSMUST00000208863.2 |
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chrX_-_161426542 | 7.80 |
ENSMUST00000101102.2
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr15_-_75438660 | 7.73 |
ENSMUST00000065417.15
|
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr7_+_16609227 | 7.72 |
ENSMUST00000108493.3
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr7_-_30826184 | 7.72 |
ENSMUST00000211945.2
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr14_-_119162736 | 7.63 |
ENSMUST00000004055.10
|
Dzip1
|
DAZ interacting protein 1 |
| chr1_+_75483721 | 7.55 |
ENSMUST00000037330.5
|
Inha
|
inhibin alpha |
| chr2_+_118507709 | 7.38 |
ENSMUST00000110853.8
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr4_+_58943574 | 7.38 |
ENSMUST00000107554.2
|
Zkscan16
|
zinc finger with KRAB and SCAN domains 16 |
| chr6_-_92511836 | 7.37 |
ENSMUST00000113446.8
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr6_-_119307685 | 7.25 |
ENSMUST00000112756.8
ENSMUST00000068351.14 |
Lrtm2
|
leucine-rich repeats and transmembrane domains 2 |
| chr2_+_118610184 | 7.23 |
ENSMUST00000063975.10
ENSMUST00000037547.9 ENSMUST00000110846.8 ENSMUST00000110843.2 |
Disp2
|
dispatched RND tramsporter family member 2 |
| chr2_-_180798785 | 7.22 |
ENSMUST00000055990.8
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr6_+_104469751 | 7.15 |
ENSMUST00000161446.2
ENSMUST00000161070.8 ENSMUST00000089215.12 |
Cntn6
|
contactin 6 |
| chr15_-_33687986 | 7.14 |
ENSMUST00000042021.5
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr18_+_34973605 | 7.12 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr17_+_55752485 | 6.96 |
ENSMUST00000025000.4
|
St6gal2
|
beta galactoside alpha 2,6 sialyltransferase 2 |
| chr7_-_4847673 | 6.96 |
ENSMUST00000066041.12
ENSMUST00000119433.4 |
Shisa7
|
shisa family member 7 |
| chrX_-_161426624 | 6.95 |
ENSMUST00000112334.8
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr18_-_43032535 | 6.94 |
ENSMUST00000120632.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr17_+_26036893 | 6.90 |
ENSMUST00000235694.2
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr6_+_118043307 | 6.82 |
ENSMUST00000203804.3
ENSMUST00000203482.2 |
Rasgef1a
|
RasGEF domain family, member 1A |
| chr1_+_75526225 | 6.74 |
ENSMUST00000154101.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr8_-_85663976 | 6.73 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr9_-_21963306 | 6.72 |
ENSMUST00000003501.9
ENSMUST00000215901.2 |
Elavl3
|
ELAV like RNA binding protein 3 |
| chr12_-_17226889 | 6.71 |
ENSMUST00000170580.3
|
Kcnf1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr5_+_35915217 | 6.70 |
ENSMUST00000101280.10
ENSMUST00000054598.12 ENSMUST00000114205.8 ENSMUST00000114206.9 |
Ablim2
|
actin-binding LIM protein 2 |
| chrX_+_134739783 | 6.70 |
ENSMUST00000173804.8
ENSMUST00000113136.8 |
Gprasp2
|
G protein-coupled receptor associated sorting protein 2 |
| chrX_+_47608122 | 6.68 |
ENSMUST00000033430.3
|
Rab33a
|
RAB33A, member RAS oncogene family |
| chr6_-_23839136 | 6.67 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr15_+_88943916 | 6.54 |
ENSMUST00000161372.2
ENSMUST00000162424.2 |
Panx2
|
pannexin 2 |
| chr2_+_14878480 | 6.48 |
ENSMUST00000114719.7
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr3_-_84212069 | 6.45 |
ENSMUST00000107692.8
|
Trim2
|
tripartite motif-containing 2 |
| chr14_+_68321302 | 6.44 |
ENSMUST00000022639.8
|
Nefl
|
neurofilament, light polypeptide |
| chr9_-_75518387 | 6.44 |
ENSMUST00000215462.2
ENSMUST00000217233.2 ENSMUST00000215614.2 |
Tmod2
|
tropomodulin 2 |
| chr13_+_97377604 | 6.43 |
ENSMUST00000041623.9
|
Enc1
|
ectodermal-neural cortex 1 |
| chr14_-_28691423 | 6.41 |
ENSMUST00000225985.2
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr2_-_27032441 | 6.40 |
ENSMUST00000151224.3
|
Fam163b
|
family with sequence similarity 163, member B |
| chr18_-_43032359 | 6.39 |
ENSMUST00000117687.8
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr9_-_20657643 | 6.33 |
ENSMUST00000215999.2
|
Olfm2
|
olfactomedin 2 |
| chr9_-_62444318 | 6.33 |
ENSMUST00000048043.12
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr12_+_108300599 | 6.27 |
ENSMUST00000021684.6
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr7_+_4122523 | 6.26 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr14_-_76794103 | 6.21 |
ENSMUST00000064517.9
ENSMUST00000228055.2 |
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr13_-_25454058 | 6.18 |
ENSMUST00000057866.13
|
Nrsn1
|
neurensin 1 |
| chr8_-_126625029 | 6.14 |
ENSMUST00000047239.13
ENSMUST00000131127.3 |
Pcnx2
|
pecanex homolog 2 |
| chrX_-_16777913 | 6.14 |
ENSMUST00000040134.8
|
Ndp
|
Norrie disease (pseudoglioma) (human) |
| chr17_-_24908874 | 6.13 |
ENSMUST00000007236.5
|
Syngr3
|
synaptogyrin 3 |
| chr19_+_45549009 | 6.03 |
ENSMUST00000047057.9
|
Gm17018
|
predicted gene 17018 |
| chr10_+_29087658 | 6.00 |
ENSMUST00000213489.2
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr5_+_35915290 | 6.00 |
ENSMUST00000114204.8
ENSMUST00000129347.8 |
Ablim2
|
actin-binding LIM protein 2 |
| chr9_-_35469818 | 6.00 |
ENSMUST00000034612.7
|
Ddx25
|
DEAD box helicase 25 |
| chrX_-_94240056 | 5.99 |
ENSMUST00000200628.2
ENSMUST00000197364.5 ENSMUST00000181987.8 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr7_-_30156826 | 5.99 |
ENSMUST00000045817.14
|
Kirrel2
|
kirre like nephrin family adhesion molecule 2 |
| chr2_+_119572770 | 5.98 |
ENSMUST00000028758.8
|
Itpka
|
inositol 1,4,5-trisphosphate 3-kinase A |
| chr10_-_80861239 | 5.97 |
ENSMUST00000055125.5
|
Diras1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr17_+_55752370 | 5.97 |
ENSMUST00000133899.2
ENSMUST00000086878.10 |
St6gal2
|
beta galactoside alpha 2,6 sialyltransferase 2 |
| chr5_+_120787253 | 5.95 |
ENSMUST00000156722.2
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr7_+_4122555 | 5.91 |
ENSMUST00000079415.12
|
Ttyh1
|
tweety family member 1 |
| chr6_-_124410452 | 5.84 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr6_-_42670021 | 5.78 |
ENSMUST00000121083.8
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr8_+_10203911 | 5.77 |
ENSMUST00000208309.2
ENSMUST00000207477.2 |
Myo16
|
myosin XVI |
| chr18_-_43032514 | 5.75 |
ENSMUST00000236238.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr6_+_104470181 | 5.74 |
ENSMUST00000162872.2
|
Cntn6
|
contactin 6 |
| chrX_-_156826262 | 5.71 |
ENSMUST00000026750.15
ENSMUST00000112513.2 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr2_-_151474391 | 5.70 |
ENSMUST00000137936.2
ENSMUST00000146172.8 ENSMUST00000094456.10 ENSMUST00000148755.8 ENSMUST00000109875.8 ENSMUST00000028951.14 ENSMUST00000109877.10 |
Snph
|
syntaphilin |
| chrX_+_133305529 | 5.69 |
ENSMUST00000113224.9
ENSMUST00000113226.2 |
Drp2
|
dystrophin related protein 2 |
| chr1_+_162466717 | 5.67 |
ENSMUST00000028020.11
|
Myoc
|
myocilin |
| chr1_-_84673903 | 5.63 |
ENSMUST00000049126.13
|
Dner
|
delta/notch-like EGF repeat containing |
| chr4_-_151192911 | 5.62 |
ENSMUST00000105670.8
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr15_+_81820954 | 5.60 |
ENSMUST00000038757.8
ENSMUST00000230633.2 |
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr10_+_29087602 | 5.59 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr1_+_158189831 | 5.57 |
ENSMUST00000193042.6
ENSMUST00000046110.16 |
Astn1
|
astrotactin 1 |
| chr11_-_115258508 | 5.56 |
ENSMUST00000044152.13
ENSMUST00000106542.9 |
Hid1
|
HID1 domain containing |
| chr6_+_47221293 | 5.54 |
ENSMUST00000199100.5
|
Cntnap2
|
contactin associated protein-like 2 |
| chr4_-_91288221 | 5.52 |
ENSMUST00000102799.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr7_+_16678568 | 5.49 |
ENSMUST00000094807.6
|
Pnmal2
|
PNMA-like 2 |
| chr14_-_78970160 | 5.47 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr14_-_55150547 | 5.47 |
ENSMUST00000228495.3
ENSMUST00000228119.3 ENSMUST00000050772.10 ENSMUST00000231305.2 |
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr1_+_66507523 | 5.47 |
ENSMUST00000061620.17
ENSMUST00000212557.3 |
Unc80
|
unc-80, NALCN activator |
| chr6_-_77956499 | 5.46 |
ENSMUST00000159626.8
ENSMUST00000075340.12 ENSMUST00000162273.2 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr4_+_130202388 | 5.37 |
ENSMUST00000070532.8
|
Fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr7_-_84059321 | 5.36 |
ENSMUST00000085077.5
ENSMUST00000207769.2 |
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr10_-_79449826 | 5.35 |
ENSMUST00000059699.9
ENSMUST00000178228.3 |
C2cd4c
|
C2 calcium-dependent domain containing 4C |
| chr2_+_102488985 | 5.35 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr16_-_31133622 | 5.35 |
ENSMUST00000115230.2
ENSMUST00000130560.8 |
Apod
|
apolipoprotein D |
| chr5_+_22951015 | 5.34 |
ENSMUST00000197992.2
|
Lhfpl3
|
lipoma HMGIC fusion partner-like 3 |
| chr4_-_68872585 | 5.31 |
ENSMUST00000030036.6
|
Brinp1
|
bone morphogenic protein/retinoic acid inducible neural specific 1 |
| chr19_-_41732104 | 5.29 |
ENSMUST00000025993.10
|
Slit1
|
slit guidance ligand 1 |
| chr6_-_115228800 | 5.28 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr3_-_152373997 | 5.28 |
ENSMUST00000045262.11
|
Ak5
|
adenylate kinase 5 |
| chr15_+_82140224 | 5.25 |
ENSMUST00000143238.2
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr2_-_163760603 | 5.25 |
ENSMUST00000044734.3
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr11_-_33942981 | 5.24 |
ENSMUST00000238903.2
|
Kcnip1
|
Kv channel-interacting protein 1 |
| chr8_-_41087793 | 5.22 |
ENSMUST00000173957.2
ENSMUST00000048898.17 ENSMUST00000174205.8 |
Mtmr7
|
myotubularin related protein 7 |
| chrX_+_142301572 | 5.20 |
ENSMUST00000033640.14
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chrX_-_94209913 | 5.17 |
ENSMUST00000113873.9
ENSMUST00000113876.9 ENSMUST00000199920.5 ENSMUST00000113885.8 ENSMUST00000113883.8 ENSMUST00000196012.2 ENSMUST00000182001.8 ENSMUST00000113878.8 ENSMUST00000113882.8 ENSMUST00000182562.2 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr2_-_127363251 | 5.16 |
ENSMUST00000028850.15
ENSMUST00000103215.11 |
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr9_+_102988940 | 5.14 |
ENSMUST00000189134.2
ENSMUST00000035155.8 |
Rab6b
|
RAB6B, member RAS oncogene family |
| chr5_+_144482693 | 5.12 |
ENSMUST00000071782.8
|
Nptx2
|
neuronal pentraxin 2 |
| chr4_+_102617332 | 5.10 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr1_-_125839897 | 5.09 |
ENSMUST00000159417.2
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr3_-_107366868 | 5.08 |
ENSMUST00000009617.10
ENSMUST00000238670.2 |
Kcnc4
|
potassium voltage gated channel, Shaw-related subfamily, member 4 |
| chr2_-_162502994 | 5.08 |
ENSMUST00000109442.8
ENSMUST00000109445.9 ENSMUST00000109443.8 ENSMUST00000109441.2 |
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr4_+_104224774 | 5.08 |
ENSMUST00000106830.9
|
Dab1
|
disabled 1 |
| chr8_+_46111703 | 5.08 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_-_115229128 | 5.07 |
ENSMUST00000032462.9
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr16_+_20514925 | 5.06 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr9_+_45029080 | 5.06 |
ENSMUST00000170998.9
ENSMUST00000093855.4 |
Scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr10_+_3316057 | 5.00 |
ENSMUST00000043374.7
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr11_-_115258493 | 4.97 |
ENSMUST00000123428.2
|
Hid1
|
HID1 domain containing |
| chr5_-_142594549 | 4.87 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr7_-_100307601 | 4.87 |
ENSMUST00000138830.2
ENSMUST00000107044.10 ENSMUST00000116287.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr17_-_25789652 | 4.86 |
ENSMUST00000025003.10
ENSMUST00000173447.2 |
Sox8
|
SRY (sex determining region Y)-box 8 |
| chr10_-_32765671 | 4.85 |
ENSMUST00000218645.2
|
Nkain2
|
Na+/K+ transporting ATPase interacting 2 |
| chr1_+_182591425 | 4.83 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr7_+_40547608 | 4.82 |
ENSMUST00000044705.12
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr7_+_63094489 | 4.80 |
ENSMUST00000058476.14
|
Otud7a
|
OTU domain containing 7A |
| chr3_+_107803225 | 4.78 |
ENSMUST00000172247.8
ENSMUST00000167387.8 |
Gstm5
|
glutathione S-transferase, mu 5 |
| chr12_+_81073573 | 4.76 |
ENSMUST00000110347.9
ENSMUST00000021564.11 ENSMUST00000129362.2 |
Smoc1
|
SPARC related modular calcium binding 1 |
| chr1_+_158189900 | 4.76 |
ENSMUST00000170718.7
|
Astn1
|
astrotactin 1 |
| chr4_+_137434767 | 4.75 |
ENSMUST00000097837.11
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr5_+_117979899 | 4.74 |
ENSMUST00000142742.9
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr5_+_27022355 | 4.73 |
ENSMUST00000071500.13
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr1_+_75362187 | 4.72 |
ENSMUST00000137868.8
|
Speg
|
SPEG complex locus |
| chr15_-_71599664 | 4.71 |
ENSMUST00000022953.10
|
Fam135b
|
family with sequence similarity 135, member B |
| chr3_+_145464413 | 4.68 |
ENSMUST00000029845.15
|
Ddah1
|
dimethylarginine dimethylaminohydrolase 1 |
| chrX_+_142447361 | 4.68 |
ENSMUST00000126592.8
ENSMUST00000156449.8 ENSMUST00000155215.8 ENSMUST00000112865.8 |
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr11_+_7013422 | 4.66 |
ENSMUST00000020706.5
|
Adcy1
|
adenylate cyclase 1 |
| chr9_-_57513510 | 4.65 |
ENSMUST00000215487.2
ENSMUST00000045068.10 |
Cplx3
|
complexin 3 |
| chr10_-_61946724 | 4.65 |
ENSMUST00000142821.8
ENSMUST00000124615.8 ENSMUST00000064050.5 ENSMUST00000125704.8 ENSMUST00000142796.8 |
Fam241b
|
family with sequence similarity 241, member B |
| chr13_+_97208069 | 4.64 |
ENSMUST00000042517.8
|
Fam169a
|
family with sequence similarity 169, member A |
| chr3_-_10505113 | 4.60 |
ENSMUST00000029047.12
ENSMUST00000195822.2 ENSMUST00000099223.11 |
Snx16
|
sorting nexin 16 |
| chr15_-_44651411 | 4.53 |
ENSMUST00000090057.6
ENSMUST00000110269.8 ENSMUST00000228639.2 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chr6_-_86374080 | 4.53 |
ENSMUST00000204116.2
ENSMUST00000153723.3 ENSMUST00000032065.15 |
Pcyox1
|
prenylcysteine oxidase 1 |
| chr6_-_23839419 | 4.48 |
ENSMUST00000115358.9
ENSMUST00000163871.9 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr14_+_32043944 | 4.48 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr6_-_114018982 | 4.48 |
ENSMUST00000101045.10
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr11_+_31950452 | 4.40 |
ENSMUST00000109409.8
ENSMUST00000020537.9 |
Nsg2
|
neuron specific gene family member 2 |
| chr3_+_27237143 | 4.39 |
ENSMUST00000091284.5
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chrX_-_132589727 | 4.38 |
ENSMUST00000149154.8
|
Pcdh19
|
protocadherin 19 |
| chr19_+_22425534 | 4.38 |
ENSMUST00000235522.2
ENSMUST00000236372.2 ENSMUST00000238066.2 ENSMUST00000235780.2 ENSMUST00000236804.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr15_-_103446354 | 4.38 |
ENSMUST00000023133.8
|
Ppp1r1a
|
protein phosphatase 1, regulatory inhibitor subunit 1A |
| chr12_+_24622274 | 4.37 |
ENSMUST00000085553.13
|
Grhl1
|
grainyhead like transcription factor 1 |
| chr3_-_127019496 | 4.36 |
ENSMUST00000182064.9
ENSMUST00000182452.8 |
Ank2
|
ankyrin 2, brain |
| chr2_-_25209199 | 4.36 |
ENSMUST00000114312.2
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr5_-_18565353 | 4.36 |
ENSMUST00000074694.7
|
Gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting 1 |
| chr8_+_85065268 | 4.34 |
ENSMUST00000238701.2
|
Cacna1a
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr1_-_134883577 | 4.33 |
ENSMUST00000168381.8
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr2_+_181356797 | 4.30 |
ENSMUST00000071585.10
ENSMUST00000238942.2 ENSMUST00000148334.8 ENSMUST00000108763.8 |
Oprl1
|
opioid receptor-like 1 |
| chr4_-_155430153 | 4.30 |
ENSMUST00000103178.11
|
Prkcz
|
protein kinase C, zeta |
| chr9_-_83688294 | 4.28 |
ENSMUST00000034796.14
ENSMUST00000183614.2 |
Elovl4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 |
| chr15_+_81695615 | 4.27 |
ENSMUST00000023024.8
|
Tef
|
thyrotroph embryonic factor |
| chr15_-_75963446 | 4.26 |
ENSMUST00000228366.3
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr2_-_180596469 | 4.25 |
ENSMUST00000148905.8
ENSMUST00000103053.10 ENSMUST00000108873.9 |
Nkain4
|
Na+/K+ transporting ATPase interacting 4 |
| chr7_+_126549692 | 4.24 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr11_-_119937970 | 4.22 |
ENSMUST00000103020.8
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr4_+_124779592 | 4.20 |
ENSMUST00000149146.2
|
Epha10
|
Eph receptor A10 |
| chr10_-_80861357 | 4.19 |
ENSMUST00000144640.2
|
Diras1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr11_+_77353218 | 4.19 |
ENSMUST00000102493.8
|
Coro6
|
coronin 6 |
| chr7_+_101060093 | 4.14 |
ENSMUST00000084894.15
|
Gm45837
|
predicted gene 45837 |
| chr1_+_158190090 | 4.10 |
ENSMUST00000194369.6
ENSMUST00000195311.6 |
Astn1
|
astrotactin 1 |
| chr11_+_67477501 | 4.10 |
ENSMUST00000108680.2
|
Gas7
|
growth arrest specific 7 |
| chr9_-_44710480 | 4.09 |
ENSMUST00000214833.2
ENSMUST00000213972.2 ENSMUST00000214431.2 ENSMUST00000213363.2 ENSMUST00000114705.9 ENSMUST00000002100.8 |
Tmem25
|
transmembrane protein 25 |
| chr9_-_86762467 | 4.07 |
ENSMUST00000074501.12
ENSMUST00000239074.2 ENSMUST00000098495.10 ENSMUST00000036347.13 ENSMUST00000074468.13 |
Snap91
|
synaptosomal-associated protein 91 |
| chr7_+_57240250 | 4.00 |
ENSMUST00000196198.5
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.9 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 4.2 | 12.5 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 2.8 | 8.5 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 2.6 | 10.4 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 2.4 | 16.6 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.3 | 15.8 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 2.2 | 6.5 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 2.1 | 6.4 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 2.1 | 10.4 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 2.0 | 8.1 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 2.0 | 6.0 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 1.9 | 11.7 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 1.9 | 7.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 1.8 | 5.5 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.8 | 5.3 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 1.7 | 5.1 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 1.7 | 5.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.7 | 3.4 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 1.6 | 8.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 1.6 | 6.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.5 | 7.5 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 1.5 | 4.4 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.5 | 7.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 1.4 | 1.4 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 1.4 | 4.3 | GO:0099578 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 1.4 | 5.7 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 1.4 | 5.6 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 1.4 | 4.2 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 1.4 | 11.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 1.4 | 8.3 | GO:0098712 | L-glutamate import across plasma membrane(GO:0098712) |
| 1.4 | 5.5 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 1.4 | 9.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 1.3 | 5.4 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.3 | 3.8 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 1.2 | 3.7 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 1.2 | 6.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 1.2 | 3.7 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 1.2 | 4.7 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 1.2 | 7.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 1.2 | 4.7 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 1.2 | 3.5 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 1.2 | 21.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 1.2 | 14.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 1.2 | 3.5 | GO:0061193 | taste bud development(GO:0061193) |
| 1.2 | 10.5 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 1.1 | 3.4 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.1 | 14.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.1 | 1.1 | GO:0072190 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) ureter urothelium development(GO:0072190) |
| 1.1 | 4.3 | GO:0021750 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) |
| 1.1 | 13.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.1 | 5.3 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 1.1 | 3.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 1.0 | 6.2 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.0 | 3.0 | GO:1904024 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 1.0 | 11.6 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 1.0 | 5.7 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 1.0 | 3.8 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 1.0 | 2.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.9 | 0.9 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.9 | 8.4 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.9 | 3.7 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.9 | 6.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.9 | 3.6 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.9 | 12.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.8 | 5.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.8 | 5.8 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.8 | 5.8 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.8 | 4.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.8 | 3.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.8 | 4.8 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.8 | 4.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.8 | 2.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.8 | 2.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.7 | 1.5 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.7 | 4.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.7 | 21.2 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.7 | 1.4 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.7 | 4.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.7 | 2.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.7 | 4.2 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.7 | 2.1 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.7 | 2.8 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.7 | 11.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.7 | 2.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.7 | 12.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.7 | 2.7 | GO:0060278 | regulation of ovulation(GO:0060278) |
| 0.7 | 2.7 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.7 | 7.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.6 | 3.9 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.6 | 0.6 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.6 | 4.5 | GO:0048840 | otolith development(GO:0048840) |
| 0.6 | 10.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.6 | 3.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.6 | 5.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.6 | 5.6 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.6 | 1.8 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.6 | 1.8 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.6 | 4.3 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.6 | 2.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.6 | 1.8 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.6 | 2.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.6 | 3.6 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.6 | 1.8 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.6 | 3.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.6 | 7.0 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.6 | 8.1 | GO:0051901 | negative regulation of appetite(GO:0032099) positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.6 | 5.8 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.6 | 4.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.6 | 2.8 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.6 | 16.8 | GO:0060384 | innervation(GO:0060384) |
| 0.6 | 1.7 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.6 | 1.7 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.5 | 6.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.5 | 4.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 1.6 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.5 | 1.6 | GO:0036118 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.5 | 4.7 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.5 | 1.6 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.5 | 2.1 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.5 | 2.6 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.5 | 1.0 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.5 | 2.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.5 | 9.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.5 | 3.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.5 | 1.5 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.5 | 2.5 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.5 | 2.5 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.5 | 6.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.5 | 2.5 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.5 | 6.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.5 | 3.9 | GO:0031179 | peptide modification(GO:0031179) |
| 0.5 | 3.9 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.5 | 3.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.5 | 4.4 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.5 | 9.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.5 | 1.4 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.5 | 3.3 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.5 | 2.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.5 | 0.9 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 15.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.5 | 22.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.5 | 1.8 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.5 | 0.5 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.4 | 1.8 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 0.4 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.4 | 0.9 | GO:0090212 | negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.4 | 3.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.4 | 9.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.4 | 1.3 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.4 | 1.3 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.4 | 2.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.4 | 1.3 | GO:1904954 | Spemann organizer formation(GO:0060061) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.4 | 2.9 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.4 | 1.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.4 | 1.2 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.4 | 3.2 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.4 | 0.8 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.4 | 3.6 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.4 | 2.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.4 | 7.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.4 | 2.4 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.4 | 1.2 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.4 | 2.4 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 | 1.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.4 | 5.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.4 | 2.0 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.4 | 6.0 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.4 | 3.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.4 | 2.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 33.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.4 | 3.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.4 | 7.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 2.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.4 | 2.9 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.4 | 2.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.4 | 5.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.4 | 1.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.4 | 4.3 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 1.4 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 1.4 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) nonassociative learning(GO:0046958) |
| 0.3 | 3.5 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.3 | 4.1 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.3 | 2.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 2.0 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 3.6 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 0.3 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.3 | 2.3 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 1.9 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.3 | 4.7 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.3 | 4.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 1.6 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.3 | 1.3 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.3 | 2.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.3 | 3.8 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.3 | 0.9 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.3 | 1.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 6.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 12.3 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.3 | 4.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 1.2 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.3 | 4.5 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 0.9 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.3 | 0.9 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.3 | 0.3 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.3 | 1.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.3 | 2.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.3 | 1.4 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 2.0 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.3 | 4.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 5.9 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.3 | 2.5 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.3 | 0.8 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) fasciculation of motor neuron axon(GO:0097156) |
| 0.3 | 0.8 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.3 | 0.8 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.3 | 0.8 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.3 | 4.6 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.3 | 9.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.3 | 7.5 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.3 | 2.9 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.3 | 2.1 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.3 | 2.1 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.3 | 2.6 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.3 | 1.1 | GO:2001107 | negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.3 | 1.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 2.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.3 | 3.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.3 | 0.8 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 3.4 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.3 | 1.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.3 | 1.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.3 | 2.5 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.2 | 2.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 2.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 4.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 7.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.2 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.2 | 1.6 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.2 | 0.9 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 0.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.6 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.2 | 4.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 1.8 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.2 | 3.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 0.2 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.2 | 2.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 7.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 2.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 1.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.2 | 0.9 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 1.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.9 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.2 | 8.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 9.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.2 | 1.9 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 3.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 2.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 7.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.2 | 1.0 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 1.6 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.2 | 1.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 2.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 3.3 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.2 | 0.8 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 2.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 9.0 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 3.6 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 1.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 2.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.2 | 8.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 0.7 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 1.3 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.2 | 1.6 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.2 | 0.5 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 0.7 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.2 | 0.9 | GO:0042748 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.2 | 1.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 4.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 1.4 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.2 | 7.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 1.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 2.4 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.2 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 1.0 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 3.8 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.2 | 13.9 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.2 | 3.6 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 3.9 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.2 | 2.9 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.2 | 8.2 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.2 | 0.5 | GO:1903027 | asymmetric Golgi ribbon formation(GO:0090164) regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.2 | 0.5 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 2.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 0.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 1.7 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 0.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 0.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 0.6 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.2 | 0.5 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) histone H3-R2 methylation(GO:0034970) |
| 0.2 | 0.6 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 6.3 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.1 | 1.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 2.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 2.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.4 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 3.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 5.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 3.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.7 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 9.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.0 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 2.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 1.0 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 1.0 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 0.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 3.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 3.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 1.8 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 2.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.5 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 2.8 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 0.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.5 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.4 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 1.3 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.1 | 1.3 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.8 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 11.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 4.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 1.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.9 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 2.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 4.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.8 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.7 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 1.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.1 | 0.9 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.9 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.8 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.1 | 0.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 2.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 6.4 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 1.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.2 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.2 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 1.5 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 5.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.6 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.6 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 3.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 2.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.3 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 2.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.9 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 4.2 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.0 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 1.9 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.1 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.1 | 1.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 1.1 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 4.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 5.3 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 8.4 | GO:0048709 | oligodendrocyte differentiation(GO:0048709) |
| 0.1 | 2.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.7 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.6 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.5 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.1 | 0.8 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.9 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 2.0 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 2.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.2 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 1.8 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.1 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 3.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 9.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 1.3 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.4 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 1.0 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 2.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 3.9 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 1.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 1.4 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 0.2 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 1.9 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 6.4 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 0.8 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.1 | 0.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.8 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 1.6 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 7.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 1.0 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.8 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.5 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 0.5 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.1 | 0.5 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.0 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 2.8 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 2.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.4 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 1.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 1.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.2 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) protein localization to nuclear pore(GO:0090204) |
| 0.1 | 3.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.1 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 3.7 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 1.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 1.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.4 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.1 | 0.2 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 0.4 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 1.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.1 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.1 | 0.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 2.2 | GO:0021879 | forebrain neuron differentiation(GO:0021879) |
| 0.1 | 0.7 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.1 | 0.6 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 1.9 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.1 | 1.0 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.6 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 1.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 1.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 3.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 5.5 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.7 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.6 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.0 | 1.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0071549 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 4.6 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 1.8 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 1.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 2.3 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.9 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.3 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.4 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.4 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 1.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 1.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 2.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 1.2 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.1 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 1.2 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 0.3 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 2.2 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 2.6 | 12.8 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 2.4 | 12.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 2.1 | 6.2 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 2.0 | 8.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 2.0 | 4.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 1.9 | 11.7 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 1.1 | 4.3 | GO:0019034 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 1.0 | 4.1 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 1.0 | 7.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.0 | 3.8 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.9 | 11.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.8 | 2.5 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.8 | 20.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.8 | 13.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.8 | 4.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.7 | 12.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.7 | 2.9 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.7 | 2.8 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.7 | 2.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.7 | 6.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.7 | 2.8 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.7 | 2.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.7 | 8.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 1.8 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.6 | 5.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.6 | 1.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.6 | 16.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.6 | 3.4 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.6 | 3.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.5 | 27.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.5 | 15.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.5 | 5.9 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.5 | 8.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.5 | 2.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 18.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 2.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.5 | 22.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.5 | 0.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.5 | 17.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.5 | 5.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.5 | 6.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.5 | 3.9 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.5 | 3.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.5 | 5.1 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.5 | 6.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 1.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 8.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 1.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.4 | 0.9 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 2.5 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.4 | 4.5 | GO:0097433 | dense body(GO:0097433) |
| 0.4 | 7.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.4 | 1.2 | GO:0097635 | Atg12-Atg5-Atg16 complex(GO:0034274) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.4 | 2.0 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.4 | 6.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 1.9 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.4 | 2.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 3.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 2.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.4 | 7.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.4 | 3.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.4 | 1.8 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.3 | 11.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 4.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.3 | 0.7 | GO:0044753 | amphisome(GO:0044753) |
| 0.3 | 10.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 2.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 3.5 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 0.9 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 0.6 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.3 | 6.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 6.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 2.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.3 | 3.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 0.8 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.3 | 29.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.3 | 13.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 4.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 8.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.3 | 3.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 1.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 2.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.3 | 4.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 2.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 1.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 4.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 2.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 6.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 6.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 5.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 68.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 2.9 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.2 | 1.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 22.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 20.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 4.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 1.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 1.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 4.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 21.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.2 | 5.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 2.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 8.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 11.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 44.1 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.2 | 3.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.2 | 1.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.9 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.2 | 1.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 0.8 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.2 | 0.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 3.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.2 | 3.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 2.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 7.2 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 30.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.9 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 6.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 1.0 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 19.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 2.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.5 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.6 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 3.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 0.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.9 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 47.4 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 2.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.7 | GO:0030681 | ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 3.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 5.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 3.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 4.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 13.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 3.1 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 4.1 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.9 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 3.0 | 12.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 2.6 | 7.8 | GO:0051424 | corticotropin-releasing hormone binding(GO:0051424) |
| 2.4 | 21.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 2.4 | 16.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 2.2 | 12.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 2.1 | 20.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 2.1 | 10.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.8 | 5.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.6 | 12.5 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.6 | 12.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 1.5 | 4.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 1.5 | 12.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.4 | 4.2 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 1.4 | 8.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 1.4 | 8.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 1.2 | 16.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 1.2 | 3.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.1 | 3.4 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 1.1 | 3.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 1.1 | 3.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 1.0 | 7.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.0 | 5.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.0 | 9.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.0 | 6.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.0 | 6.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.0 | 3.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.0 | 10.5 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.9 | 6.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.9 | 3.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.9 | 5.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.9 | 5.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.9 | 12.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.8 | 2.5 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.8 | 4.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.8 | 28.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.7 | 3.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.7 | 3.7 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.7 | 17.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.7 | 2.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.7 | 2.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.7 | 2.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.7 | 6.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.7 | 15.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.7 | 3.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.7 | 2.7 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.7 | 4.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.7 | 4.6 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.7 | 3.9 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.7 | 8.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.7 | 6.5 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.6 | 3.9 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.6 | 3.9 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.6 | 5.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.6 | 3.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 1.9 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.6 | 13.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 3.7 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.6 | 4.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.6 | 4.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.6 | 1.8 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.6 | 3.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.6 | 2.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.6 | 10.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.6 | 6.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.6 | 1.7 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.6 | 3.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.6 | 12.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.6 | 2.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.5 | 4.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 4.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 3.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.5 | 7.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.5 | 3.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.5 | 1.6 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.5 | 2.5 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.5 | 1.5 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.5 | 4.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.5 | 9.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.5 | 1.9 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.5 | 1.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.4 | 3.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 4.4 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.4 | 5.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.4 | 1.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.4 | 2.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 1.7 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.4 | 1.3 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.4 | 4.3 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.4 | 4.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 3.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.4 | 2.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.4 | 11.7 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.4 | 2.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.4 | 18.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.4 | 19.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.4 | 9.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 3.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.4 | 4.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.4 | 1.5 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.4 | 9.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 4.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.4 | 2.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 5.8 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.4 | 1.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.4 | 1.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.4 | 4.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 5.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.4 | 3.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 2.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.3 | 1.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 4.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 5.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 1.0 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.3 | 8.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 4.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 1.3 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.3 | 1.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 1.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 0.9 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.3 | 2.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 2.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 1.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 0.9 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 8.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 4.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 5.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 1.1 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 9.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 8.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 4.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 0.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.3 | 2.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.3 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.3 | 4.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 2.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 1.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 2.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 30.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 17.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 2.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 1.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 4.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 17.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 7.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 3.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 16.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 2.5 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.2 | 1.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.7 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 1.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 3.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.2 | 1.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.8 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.2 | 3.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 5.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.8 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 1.6 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 0.8 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.2 | 1.9 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 0.9 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 1.3 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.2 | 3.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 0.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 0.5 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.2 | 2.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 0.5 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 0.9 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 1.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 8.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 8.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 9.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 0.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 0.3 | GO:0033265 | choline binding(GO:0033265) |
| 0.2 | 6.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 1.9 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.2 | 3.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 0.8 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.2 | 2.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 0.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 2.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 1.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.4 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 4.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 9.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 4.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 4.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 4.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 3.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 15.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.6 | GO:0008443 | 6-phosphofructokinase activity(GO:0003872) phosphofructokinase activity(GO:0008443) |
| 0.1 | 0.7 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 2.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 5.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 7.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 4.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.1 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.8 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.1 | 2.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.0 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 1.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.7 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 1.4 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 1.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 2.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.7 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.9 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 4.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.2 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 2.7 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 4.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 1.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 5.6 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 18.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 1.0 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 2.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 3.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.3 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.1 | 0.2 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 6.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.4 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 1.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 5.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.2 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 7.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 2.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 3.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 2.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 3.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 12.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 2.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 8.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.3 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 1.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 3.8 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.9 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.0 | 0.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 2.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.9 | 2.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.5 | 25.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 3.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 17.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 3.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 9.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 14.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 9.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 6.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 11.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 6.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 2.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 22.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 2.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 6.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 3.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 20.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 27.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 5.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 1.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 7.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 3.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 3.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 0.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 3.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 22.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 0.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 0.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 31.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.8 | 27.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.8 | 9.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.8 | 21.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.8 | 10.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.7 | 4.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.7 | 8.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.5 | 28.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 3.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.5 | 10.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.4 | 8.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 25.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 9.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.4 | 6.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 6.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.4 | 6.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 1.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 3.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 3.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 3.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 4.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 12.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 6.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 8.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 4.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 3.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 3.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 8.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 7.3 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.2 | 27.0 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.2 | 5.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 5.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 1.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 3.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 10.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 8.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 5.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 3.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 5.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 4.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 5.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 4.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 4.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 2.3 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 8.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 8.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 4.0 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |
| 0.1 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 9.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 4.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |